"VSports注册入口" Same Exposure but Two Radically Different Responses to Antibiotics: Resilience of the Salivary Microbiome versus Long-Term Microbial Shifts in Feces
- PMID: 26556275
- PMCID: PMC4659469 (V体育官网入口)
- DOI: 10.1128/mBio.01693-15
Same Exposure but Two Radically Different Responses to Antibiotics: Resilience of the Salivary Microbiome versus Long-Term Microbial Shifts in Feces
Abstract
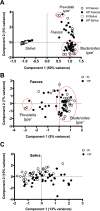
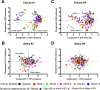
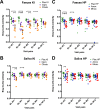
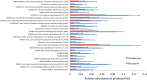
Due to the spread of resistance, antibiotic exposure receives increasing attention. Ecological consequences for the different niches of individual microbiomes are, however, largely ignored. Here, we report the effects of widely used antibiotics (clindamycin, ciprofloxacin, amoxicillin, and minocycline) with different modes of action on the ecology of both the gut and the oral microbiomes in 66 healthy adults from the United Kingdom and Sweden in a two-center randomized placebo-controlled clinical trial. Feces and saliva were collected at baseline, immediately after exposure, and 1, 2, 4, and 12 months after administration of antibiotics or placebo. Sequences of 16S rRNA gene amplicons from all samples and metagenomic shotgun sequences from selected baseline and post-antibiotic-treatment sample pairs were analyzed VSports手机版. Additionally, metagenomic predictions based on 16S rRNA gene amplicon data were performed using PICRUSt. The salivary microbiome was found to be significantly more robust, whereas the antibiotics negatively affected the fecal microbiome: in particular, health-associated butyrate-producing species became strongly underrepresented. Additionally, exposure to different antibiotics enriched genes associated with antibiotic resistance. In conclusion, healthy individuals, exposed to a single antibiotic treatment, undergo considerable microbial shifts and enrichment in antibiotic resistance in their feces, while their salivary microbiome composition remains unexpectedly stable. The health-related consequences for the gut microbiome should increase the awareness of the individual risks involved with antibiotic use, especially in a (diseased) population with an already dysregulated microbiome. On the other hand, understanding the mechanisms behind the resilience of the oral microbiome toward ecological collapse might prove useful in combating microbial dysbiosis elsewhere in the body. .
Importance: Many health care professionals use antibiotic prophylaxis strategies to prevent infection after surgery. This practice is under debate since it enhances the spread of antibiotic resistance. Another important reason to avoid nonessential use of antibiotics, the impact on our microbiome, has hardly received attention. In this study, we assessed the impact of antibiotics on the human microbial ecology at two niches. We followed the oral and gut microbiomes in 66 individuals from before, immediately after, and up to 12 months after exposure to different antibiotic classes. The salivary microbiome recovered quickly and was surprisingly robust toward antibiotic-induced disturbance V体育安卓版. The fecal microbiome was severely affected by most antibiotics: for months, health-associated butyrate-producing species became strongly underrepresented. Additionally, there was an enrichment of genes associated with antibiotic resistance. Clearly, even a single antibiotic treatment in healthy individuals contributes to the risk of resistance development and leads to long-lasting detrimental shifts in the gut microbiome. .
Copyright © 2015 Zaura et al.
"V体育平台登录" Figures

References
-
- Cantas L, Shah SQA, Cavaco LM, Manaia CM, Walsh F, Popowska M, Garelick H, Bürgmann H, Sørum H. 2013. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front Microbiol 4:96. doi:10.3389/fmicb.2013.00096. - "V体育安卓版" DOI - PMC - PubMed
-
- Alanis AJ. 2005. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res 36:697–705. doi:10.1016/j.arcmed.2005.06.009. - VSports在线直播 - DOI - PubMed
-
- Modi SR, Collins JJ, Relman DA. 2014. Antibiotics and the gut microbiota. J Clin Invest 124:4212–4218. doi:10.1172/JCI72333. - "V体育官网入口" DOI - PMC - PubMed
Publication types
- V体育安卓版 - Actions
MeSH terms
- Actions (V体育官网入口)
- "VSports手机版" Actions
- "VSports注册入口" Actions
- V体育安卓版 - Actions
- "VSports注册入口" Actions
VSports最新版本 - Substances
- Actions (VSports最新版本)
VSports注册入口 - LinkOut - more resources
Full Text Sources
Medical
"VSports在线直播" Molecular Biology Databases

