VSports在线直播 - The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast
- PMID: 26518661
- PMCID: VSports - PMC4687421
- DOI: 10.15252/embr.201541214
VSports在线直播 - The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast
"VSports最新版本" Abstract
The maintenance of open and repressed chromatin states is crucial for the regulation of gene expression. To study the genes involved in maintaining chromatin states, we generated a random mutant library in Schizosaccharomyces pombe and monitored the silencing of reporter genes inserted into the euchromatic region adjacent to the heterochromatic mating type locus. We show that Leo1-Paf1 [a subcomplex of the RNA polymerase II-associated factor 1 complex (Paf1C)] is required to prevent the spreading of heterochromatin into euchromatin by mapping the heterochromatin mark H3K9me2 using high-resolution genomewide ChIP (ChIP-exo). Loss of Leo1-Paf1 increases heterochromatin stability at several facultative heterochromatin loci in an RNAi-independent manner VSports手机版. Instead, deletion of Leo1 decreases nucleosome turnover, leading to heterochromatin stabilization. Our data reveal that Leo1-Paf1 promotes chromatin state fluctuations by enhancing histone turnover. .
Keywords: Epe1; Paf1C; heterochromatin; histone turnover; transcription V体育安卓版. .
© 2015 The Authors. Published under the terms of the CC BY 4. 0 license. V体育ios版.
Figures

Scheme of the mating type region in Schizosaccharomyces pombe. Integration positions of reporter genes (ura4 + or ade6 +) are indicated V体育平台登录.
Spotting assay using strains with ura4 + and ade6 + integrated as in (A) on plates with added adenine (
YEA ), with low adenine (YE ), or with 5‐fluorotic acid (5FOA ).ChIP–qPCR analysis of H3K9me2 levels at the ura4 + or ade6 + insertion sites depicted in (A). ChIP was performed in three independent experiments (n = 3); error bars indicate the standard deviation (SD) V体育官网入口.
Schematic representation of the protein domains of Leo1; Sp: S VSports在线直播. pombe; Hs: human. The different colors represent conserved domains: orange: Paf1 interaction; green: conserved, potentially H3 interacting; pink: conserved region of unknown function, potentially RNA binding.
Growth curves of
WT cells and cells with different leo1 + alleles (as indicated).Bright‐field microscopy images of the indicated strains.
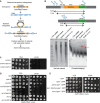
- A
Cartoon of the Hermes transposon screen.
- B
Southern blot of the Hermes strain. Probes were complementary to the kanMX region (top panel) present in both leo1::Hermes and leo1∆, leading to a band of length 7–8 kb (Hermes‐kanMX, lower panel, upper arrow) and a band of length 4–5 kb (leo1∆::kanMX) (lower panel, lower arrow) VSports app下载.
- C–E
The Ddb1 and epe1::
GFP effect on other components of Paf1C. Genetic interaction between ddb1 and cdc73 and tpr1. (C) Spotting assay with ura4 + at its endogenous locus. (D) Spotting assay with strains in ddb1∆ background. (E) Spotting assay in epe1 + ::GFP ‐derived strains.

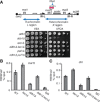
- A
Model of Paf1C with its subunits and interactors.
- B
Spotting assay using four strains with ura4 + and ade6 + integrated as in Fig 1A, carrying deletions of genes encoding different subunits of the Paf1 complex (as indicated).
- C
Position variegation effect by Leo1 and Paf1. Spotting assay of strains regrown from –Ura or 5
FOA plates with the relevant genotypes indicated. - D
Immunoblot of H2Bub1 and H4 levels in
WT , htb1‐K119R, and leo1∆ cells. Numbers indicate the H2Bub1/H4 ratio for each lane. - E, F
Growth assay performed on a low‐adenine plate (
YE ) (E) and a 5FOA ‐containing plate (F) using mutants with ura4 + or ade6 + integrated as in Fig 1A. The mutants in this panel lack different factors in the H3K4me (set1∆) or H2Bub1 (rfp1∆, shf1∆, or htb1‐K119R) pathways.
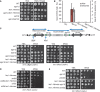
Scheme of the mat locus in S. pombe. Integration position of ura4 + is indicated (upper panel). Spotting assay on
YEA and 5FOA plates (lower panel).Ch
IP –qPCR across the XbaI site in the matK domain.Ch
IP –qPCR across the pericentric dh repeat of centromere 1.

A browser view of Ch
IP –exo signals in theWT and leo1∆ strains using an antibody against H3K9me2 showing the constitutive heterochromatin domains of chromosome 2 (indicated in red in the lower panel). The ratios ofRPKM (reads per kilobase per million mapped reads) over the indicated region are shown. Two independent experiments were performed.Browser views of H3K9me2 Ch
IP –exo signals inWT and leo1∆ cells at five facultative heterochromatin islands. TheRPKM values are shown for each gene and strain.Ch
IP –qPCR of H3K9me2 at Tf2s.Ch
IP –qPCR of Swi6 inWT , leo1∆, and leo1::Hermes strains at four loci.Spotting assay of
WT and leo1∆ with ura4 + integrated in the Tf2‐3 locus. The top row shows aWT strain with functional endogenous ura4 +; all other strains have the non‐functional ura4‐D18 allele.Spotting assay using strains with ura4 + integrated close to the
SPAC 23H3.14 locus.Ch
IP –PCR of H3K9me2 at theSPAC 23H3.14 locus. Numbers indicate ratios for the signalSPAC 23H13.14/leu1.

Ch
IP –exo for H3K9me2 at Tf2.Position variegation at Tf2 effect by leo1∆. Spotting assay on –Ura and
FOA plates of strains previously grown on –Ura orFOA plates.Spotting assay of indicated strains on
YEA and 5FOA media with ura4 + integrated at the left pericentric region of chromosome 1.Spotting assay of indicated strains on
YEA and 5FOA media with ura4 + integrated at the right pericentric region of chromosome 1.
Spotting assay to test
TBZ sensitivity of the indicated strains. Tenfold serial dilutions of the indicated cultures were grown on rich medium (YEA ) in the presence (17 μg/ml) or absence ofTBZ .Loss of minichromosome Ch16. Two independent experiments were performed; error bars show the range.
Representation of centromere 1 in S. pombe. Integration sites of ura4 + are indicated with a blue vertical arrow (upper panel). Spotting assay of strains with the relevant genotypes indicated (lower panel).
Spotting of strains lacking
HP 1 (swi6∆) orSHREC (clr3) with ura4 + integrated at the imr1.Spotting of strains lacking
FACT (pob3∆) with ura4 + integrated at the imr1.

Pie charts illustrating proportions between different
sRNA populations inWT and leo1∆.sRNA s mapped across centromere 3. The number of reads from two independent experiments was normalized to the reads mapping totRNA s. Red dots in the upper panel indicate positions oftRNA s.Growth of strains with ade6 + integrated at
IR ‐L on aYE (low‐adenine) plate.
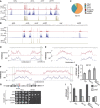
- A
sRNA distribution at tel1L and the centromere of chromosome 1. - B
sRNA distribution at tel2L, the centromere of chromosome 2, the mat region, and tel2R. - C
Distribution of
sRNA populations in ago1∆. - D–F
H3K9me2 Ch
IP –exo over centromere 1 (D), centromere 2 (E), and centromere 3 (F). Green boxes indicate the location of theBORDERLINE transcript 2013 and theIRC 3 elements 2008. - G
Spotting of strains with ura4 + at the euchromatic (inner centromeric) side of
tRNA barrier at cen1R. - H, I
H3K9me2 Ch
IP –qPCR over theIRC 1R locus (H) and the outer repeats (otr2) and central core (cnt2) of centromere 2 (I). Experiments were done in three independent experiments; error bars show theSD .

- A, B
Retrotransposable element Tf2 is silenced by an si
RNA ‐independent mechanism in leo1∆. (A) Growth of indicated strains, with ade6 + integrated at two locations, on a low‐adenine plate. (B) Distributions ofsRNA mapping to the 13 Tf2 retrotransposons. Normalized reads are aligned relative to the transcription start site, sense (top panel) and anti‐sense direction (bottom panel) inWT (blue), leo1∆ (red), and rrp6∆ (green) cells. - C, D
Histone turnover and the effects of acetyltransferases on heterochromatin formation. (C) Ch
IP –qPCR using antibody againstHA in cells with theRITE construct, using primers for dh1, tf2, matK, rad50 +, and spd1 +. ChIP was performed in two independent experiments. (D) Position variegation effect by Mst2 and Paf1. Spotting assay of strains preselected as being Ura+ orFOAR as indicated. - E
Spotting assay of strains with ura4 + integrated at matL, and ade6 + at
IR ‐L. - F
Spotting assay of strains with ura4 + integrated at matK.
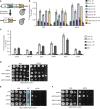
Diagram of the
RITE system. Green triangles represent LoxP sites.Ch
IP of H3‐T7 at silent chromatin regions.Ch
IP of H3‐T7 at expressed euchromatic regions transcribed byRNAPII (IRC 1R, spd1 +, rad50 +, pyk1 +, and scm3 +) orRNAPIII (tRNA ).Spotting assay using strains with ura4 + at the matK region.
Spotting assay using strains with ade6 + integrated at the
IR ‐L and ura4 + integrated at matL domains.Spotting assay using strains with ura4 + at the matK region.
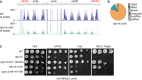
sRNA profile over centromere 3. The number of reads from two independent experiments was normalized to reads mapping totRNA s. Red dots in top panel indicate positions oftRNA s.sRNA profile in htb1‐K119R.Spotting assay of strains with ura4 + integrated at imr1R.
References
-
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 - PubMed
-
- Valenzuela L, Kamakaka RT (2006) Chromatin insulators. Annu Rev Genet 40: 107–138 - PubMed
-
- Aygun O, Mehta S, Grewal SI (2013) HDAC‐mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol 20: 547–554 - "VSports手机版" PMC - PubMed
-
- Svensson JP, Audergon P, Menendez‐Benito V, Shukla M, Sinha I, Norman‐Axelsson U, Allshire RC, Ekwall K (2015) A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin. Genome Res 25: 872–883 - "VSports在线直播" PMC - PubMed
V体育平台登录 - Publication types
- "VSports" Actions
MeSH terms
- "VSports app下载" Actions
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- Actions (VSports最新版本)
- V体育官网 - Actions
- "V体育ios版" Actions
Substances
- "VSports注册入口" Actions
- "V体育官网入口" Actions
- "VSports" Actions
Associated data
- "VSports app下载" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育官网入口)
Molecular Biology Databases

