"VSports app下载" Loss of ATRX Suppresses Resolution of Telomere Cohesion to Control Recombination in ALT Cancer Cells
- PMID: 26373281
- PMCID: PMC4573400 (V体育官网)
- DOI: 10.1016/j.ccell.2015.08.003
"V体育安卓版" Loss of ATRX Suppresses Resolution of Telomere Cohesion to Control Recombination in ALT Cancer Cells
Abstract
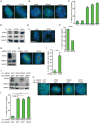
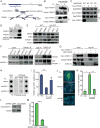
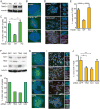
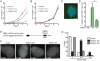
The chromatin-remodeler ATRX is frequently lost in cancer cells that use ALT (alternative lengthening of telomeres) for telomere maintenance, but its function in telomere recombination is unknown. Here we show that loss of ATRX suppresses the timely resolution of sister telomere cohesion that normally occurs prior to mitosis. In the absence of ATRX, the histone variant macroH2A1. 1 binds to the poly(ADP-ribose) polymerase tankyrase 1, preventing it from localizing to telomeres and resolving cohesion. The resulting persistent telomere cohesion promotes recombination between sister telomeres, while it suppresses inappropriate recombination between non-sisters. Forced resolution of sister telomere cohesion induces excessive recombination between non-homologs, genomic instability, and impaired cell growth, indicating the ATRX-macroH2A1. 1-tankyrase axis as a potential therapeutic target in ALT tumors VSports手机版. .
Copyright © 2015 Elsevier Inc V体育安卓版. All rights reserved. .
Figures

Comment in (V体育官网)
-
VSports在线直播 - Keeping It in the Family: ATRX Loss Promotes Persistent Sister Telomere Cohesion in ALT Cancer Cells.Cancer Cell. 2015 Sep 14;28(3):277-9. doi: 10.1016/j.ccell.2015.08.005. Cancer Cell. 2015. PMID: 26373274
"VSports最新版本" References
-
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. - PubMed
-
- Bechter OE, Zou Y, Shay JW, Wright WE. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138–1143. - "V体育平台登录" PMC - PubMed
-
- Bisht KK, Daniloski Z, Smith S. SA1 binds directly to DNA through its unique AT-hook to promote sister chromatid cohesion at telomeres. J Cell Sci. 2013;126:3493–3503. - PMC (V体育平台登录) - PubMed
-
- Bower K, Napier CE, Cole SL, Dagg RA, Lau LM, Duncan EL, Moy EL, Reddel RR. Loss of wild-type ATRX expression in somatic cell hybrids segregates with activation of Alternative Lengthening of Telomeres. PloS one. 2012;7:e50062. - V体育ios版 - PMC - PubMed
Publication types
- "V体育安卓版" Actions
MeSH terms
- Actions (V体育2025版)
- "V体育ios版" Actions
- VSports - Actions
- Actions (VSports)
- Actions (VSports)
- VSports手机版 - Actions
- "V体育平台登录" Actions
- V体育2025版 - Actions
- V体育官网入口 - Actions
- Actions (V体育官网入口)
Substances
- Actions (VSports app下载)
- "V体育官网入口" Actions
- Actions (V体育ios版)
- "V体育官网" Actions
Grants and funding
LinkOut - more resources
Full Text Sources (V体育安卓版)
"V体育安卓版" Other Literature Sources
"V体育平台登录" Molecular Biology Databases
V体育官网 - Miscellaneous

