Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells
- PMID: 26365896
- PMCID: PMC4717845
- DOI: 10.1016/j.molonc.2015.08.003
VSports最新版本 - Resistance to BRAF inhibitors induces glutamine dependency in melanoma cells
V体育安卓版 - Abstract
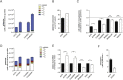
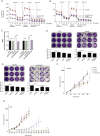
BRAF inhibitors can extend progression-free and overall survival in melanoma patients whose tumors harbor mutations in BRAF. However, the majority of patients eventually develop resistance to these drugs. Here we show that BRAF mutant melanoma cells that have developed acquired resistance to BRAF inhibitors display increased oxidative metabolism and increased dependency on mitochondria for survival. Intriguingly, the increased oxidative metabolism is associated with a switch from glucose to glutamine metabolism and an increased dependence on glutamine over glucose for proliferation. We show that the resistant cells are more sensitive to mitochondrial poisons and to inhibitors of glutaminolysis, suggesting that targeting specific metabolic pathways may offer exciting therapeutic opportunities to treat resistant tumors, or to delay emergence of resistance in the first-line setting. VSports手机版.
Keywords: BRAF; Glutaminolysis; Melanoma; Metabolism; Resistance V体育安卓版. .
Copyright © 2015 The Authors. Published by Elsevier B V体育ios版. V. All rights reserved. .
Figures


"V体育官网" References
-
- Chaneton, B. , Hillmann, P. , Zheng, L. , Martin, A.C. , Maddocks, O.D. , Chokkathukalam, A. , Coyle, J.E. , Jankevics, A. , Holding, F.P. , Vousden, K.H. , Frezza, C. , O'Reilly, M. , Gottlieb, E. , 2012. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 491, 458–462. - VSports在线直播 - PMC - PubMed
-
- Corazao-Rozas, P. , Guerreschi, P. , Jendoubi, M. , Andre, F. , Jonneaux, A. , Scalbert, C. , Garcon, G. , Malet-Martino, M. , Balayssac, S. , Rocchi, S. , Savina, A. , Formstecher, P. , Mortier, L. , Kluza, J. , Marchetti, P. , 2013. Mitochondrial oxidative stress is the Achille's heel of melanoma cells resistant to Braf-mutant inhibitor. Oncotarget. 4, 1986–1998. - PMC - PubMed
-
- Dang, L. , White, D.W. , Gross, S. , Bennett, B.D. , Bittinger, M.A. , Driggers, E.M. , Fantin, V.R. , Jang, H.G. , Jin, S. , Keenan, M.C. , Marks, K.M. , Prins, R.M. , Ward, P.S. , Yen, K.E. , Liau, L.M. , Rabinowitz, J.D. , Cantley, L.C. , Thompson, C.B. , Vander Heiden, M.G. , Su, S.M. , 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 462, 739–744. - PMC - PubMed
-
- Davies, H. , Bignell, G.R. , Cox, C. , Stephens, P. , Edkins, S. , Clegg, S. , Teague, J. , Woffendin, H. , Garnett, M.J. , Bottomley, W. , Davis, N. , Dicks, E. , Ewing, R. , Floyd, Y. , Gray, K. , Hall, S. , Hawes, R. , Hughes, J. , Kosmidou, V. , Menzies, A. , Mould, C. , Parker, A. , Stevens, C. , Watt, S. , Hooper, S. , Wilson, R. , Jayatilake, H. , Gusterson, B.A. , Cooper, C. , Shipley, J. , Hargrave, D. , Pritchard-Jones, K. , Maitland, N. , Chenevix-Trench, G. , Riggins, G.J. , Bigner, D.D. , Palmieri, G. , Cossu, A. , Flanagan, A. , Nicholson, A. , Ho, J.W. , Leung, S.Y. , Yuen, S.T. , Weber, B.L. , Seigler, H.F. , Darrow, T.L. , Paterson, H. , Marais, R. , Marshall, C.J. , Wooster, R. , Stratton, M.R. , Futreal, P.A. , 2002. Mutations of the BRAF gene in human cancer. Nature. 417, 949–954. - "V体育官网" PubMed
Publication types
"VSports手机版" MeSH terms
- VSports app下载 - Actions
- "VSports" Actions
- "VSports最新版本" Actions
- Actions (V体育官网)
Substances
- Actions (V体育官网)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
"VSports手机版" Research Materials

