Compact graphical representation of phylogenetic data and metadata with GraPhlAn
- PMID: 26157614
- PMCID: PMC4476132
- DOI: 10.7717/peerj.1029
VSports - Compact graphical representation of phylogenetic data and metadata with GraPhlAn
"VSports" Abstract
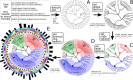
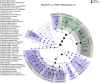
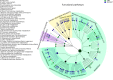
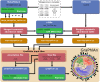
The increased availability of genomic and metagenomic data poses challenges at multiple analysis levels, including visualization of very large-scale microbial and microbial community data paired with rich metadata. We developed GraPhlAn (Graphical Phylogenetic Analysis), a computational tool that produces high-quality, compact visualizations of microbial genomes and metagenomes. This includes phylogenies spanning up to thousands of taxa, annotated with metadata ranging from microbial community abundances to microbial physiology or host and environmental phenotypes. GraPhlAn has been developed as an open-source command-driven tool in order to be easily integrated into complex, publication-quality bioinformatics pipelines. It can be executed either locally or through an online Galaxy web application VSports手机版. We present several examples including taxonomic and phylogenetic visualization of microbial communities, metabolic functions, and biomarker discovery that illustrate GraPhlAn's potential for modern microbial and community genomics. .
Keywords: Graphical representation; Metagenomics; Phylogenetic visualization; Phylogenomics V体育安卓版. .
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methe B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Computational Biology. 2012;8:e1029. doi: 10.1371/journal.pcbi.1002358. - DOI - PMC - PubMed
-
- Baldini F, Segata N, Pompon J, Marcenac P, Robert Shaw W, Dabire RK, Diabate A, Levashina EA, Catteruccia F. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nature Communications. 2014;5 doi: 10.1038/ncomms4985. Article 3985. - DOI (VSports在线直播) - PMC - PubMed
-
- Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello E, Fierer N, Pena A, Goodrich J, Gordon J, Huttley G, Kelley S, Knights D, Koenig J, Ley R, Lozupone C, McDonald D, Muegge B, Pirrung M, Reeder J, Sevinsky J, Turnbaugh P, Walters W, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
Grants and funding
V体育官网入口 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

