Widespread intron retention diversifies most cancer transcriptomes
- PMID: 26113877
- PMCID: PMC4480902
- DOI: 10.1186/s13073-015-0168-9
Widespread intron retention diversifies most cancer transcriptomes
Abstract
Background: Somatic mutations affecting components of the RNA splicing machinery occur with high frequencies across many tumor types. These mutations give rise to distinct alterations in normal splice site and exon recognition, such as unusual 3' splice site preferences, that likely contribute to tumorigenesis. VSports手机版.
Methods: We analyzed genome-wide patterns of RNA splicing across 805 matched tumor and normal control samples from 16 distinct cancer types to identify signals of abnormal cancer-associated splicing. V体育安卓版.
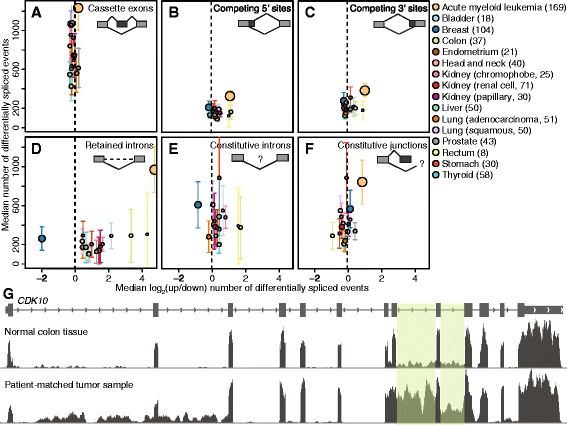
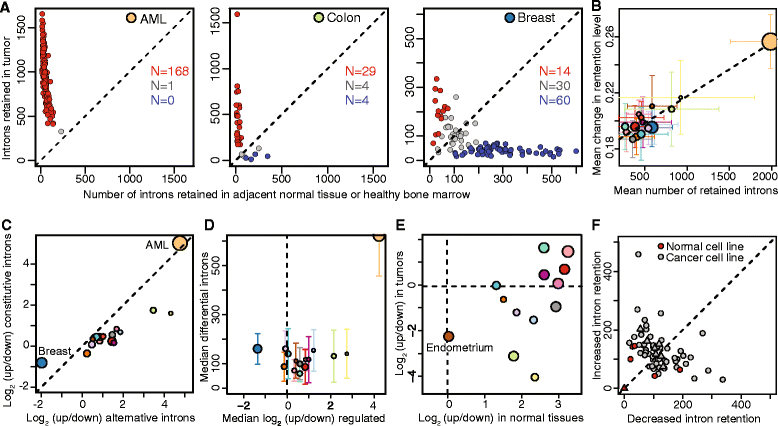
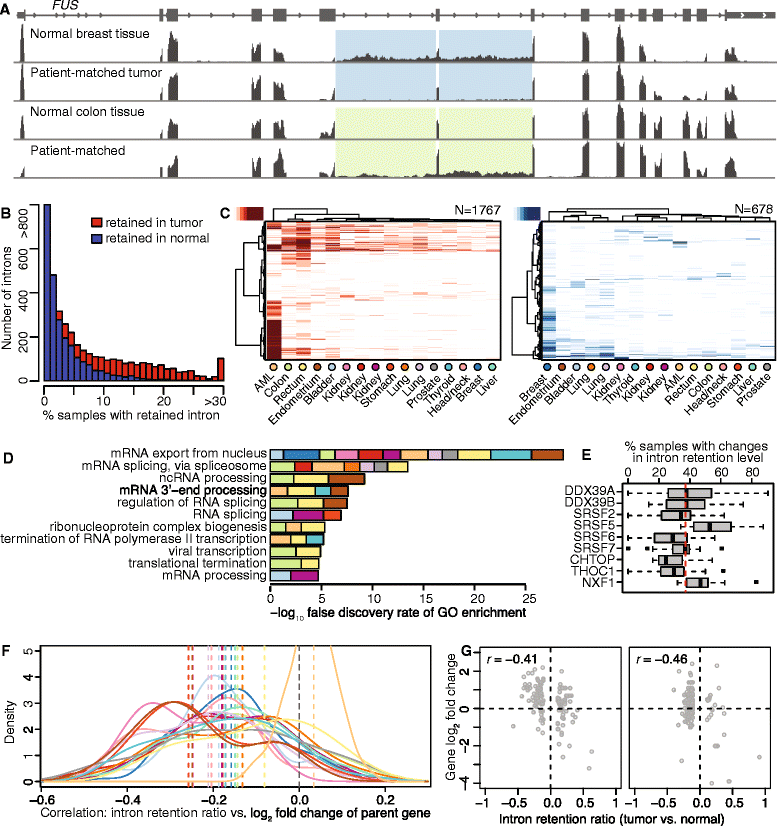
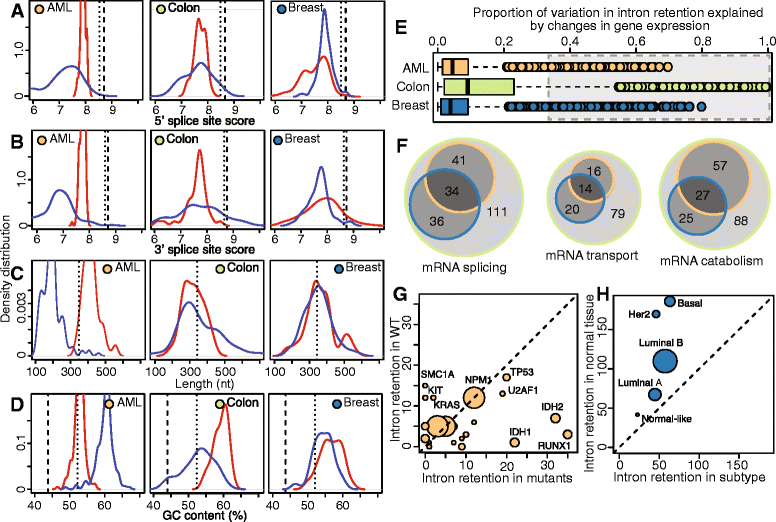
Results: We found that abnormal RNA splicing, typified by widespread intron retention, is common across cancers even in the absence of mutations directly affecting the RNA splicing machinery V体育ios版. Almost all liquid and solid cancer types exhibited frequent retention of both alternative and constitutive introns relative to control normal tissues. The sole exception was breast cancer, where intron retention typified adjacent normal rather than cancer tissue. Different introns were preferentially retained in specific cancer types, although a small subset of introns enriched for genes encoding RNA splicing and export factors exhibited frequent retention across diverse cancers. The extent of intron retention correlated with the presence of IDH1 and IDH2 mutations in acute myeloid leukemia and across molecular subtypes in breast cancer. Many introns that were preferentially retained in primary cancers were present at high levels in the cytoplasmic mRNA pools of cancer cell lines. .
Conclusions: Our data indicate that abnormal RNA splicing is a common characteristic of cancers even in the absence of mutational insults to the splicing machinery, and suggest that intron-containing mRNAs contribute to the transcriptional diversity of many cancers VSports最新版本. .
Figures
References
Grants and funding
LinkOut - more resources
VSports手机版 - Full Text Sources
"VSports手机版" Other Literature Sources
"VSports手机版" Molecular Biology Databases
Miscellaneous

