Atrx promotes heterochromatin formation at retrotransposons
- PMID: 26012739
- PMCID: VSports最新版本 - PMC4515123
- DOI: 10.15252/embr.201439937
Atrx promotes heterochromatin formation at retrotransposons (VSports注册入口)
Abstract
More than 50% of mammalian genomes consist of retrotransposon sequences. Silencing of retrotransposons by heterochromatin is essential to ensure genomic stability and transcriptional integrity. Here, we identified a short sequence element in intracisternal A particle (IAP) retrotransposons that is sufficient to trigger heterochromatin formation. We used this sequence in a genome-wide shRNA screen and identified the chromatin remodeler Atrx as a novel regulator of IAP silencing. Atrx binds to IAP elements and is necessary for efficient heterochromatin formation. In addition, Atrx facilitates a robust and largely inaccessible heterochromatin structure as Atrx knockout cells display increased chromatin accessibility at retrotransposons and non-repetitive heterochromatic loci. In summary, we demonstrate a direct role of Atrx in the establishment and robust maintenance of heterochromatin VSports手机版. .
Keywords: Atrx; Daxx; IAP retrotransposons; heterochromatin; histone H3 V体育安卓版. 3. .
© 2015 The Authors.
"VSports" Figures

Reporter assay for retrotransposon silencing. Mouse ES cells were transduced with indicated lentiviral reporter vectors, and EGFP fluorescence was monitored 2 days later by FACS. PE was used as empty channel VSports最新版本. RBS, repressor binding site of the MLV virus; IAP-GAG, GAG coding sequence of IAP retrotransposons. .
The GAG coding region of IAP retrotransposons leads to strong reporter silencing in mouse ES cells. Structure of IAP elements: LTR, long terminal repeats; UTR, 5′ untranslated region; GAG, GAG protein coding sequence; PRO, coding sequence of retroviral protease; and POL, coding sequence of retroviral polymerase. Indicated sequence elements of IAP retrotransposons were cloned upstream of the EGFP reporter cassette and tested for silencing activity in mouse ES cells and MEFs. Fold repression was calculated relative to the control EGFP vector at the same multiplicity of infection (MOI). Bar plots indicate the mean of four to seven biological replicates V体育平台登录. Error bars indicate the standard deviation. .
Identification of the 160-bp SHIN sequence. Sub-fragments of the GAG coding region were tested for silencing activity as in (B). The smallest fragment which was still able to induce full repression was designated SHIN. Bar plots indicate the mean of two to six biological replicates. Error bars indicate the standard deviation VSports注册入口. .
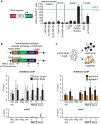
SHIN silencing depends on the Trim28/Setdb1 pathway. Mouse ES cells were transduced with the SHIN reporter or control vector, and EGFP fluorescence was monitored 2 days later by FACS. The percentage of EGFP-positive cells after SHIN reporter transduction is shown relative to the control transduction (relative % EGFP+ cells). Setdb1 knockout cells were generated by the transduction of Setdb1flox/flox ES cells with Cre-expressing virus (Cre). Trim28 knockdown was performed by the transduction of a lentiviral shRNA vector (shTrim28 #1). Bar plots indicate the mean of three to six biological replicates. Error bars indicate the standard deviation V体育2025版. .
The SHIN sequence recruits H3K9me3. Recombination-mediated cassette exchange of the indicated SHIN reporter and control vector resulted in ES cell lines HA36::SHIN and HA36::control, respectively. ChIP-qPCR analysis of H3K9me3 across the RMCE locus in HA36::SHIN and HA36::control cells. RMCE PCR amplicons (1–3) are indicated. Control regions: maj sat, major satellite repeats; pos, Polrmt; neg, Tia1; and IAP, endogenous IAP elements. N/A = sequence not present in the HA36::control construct. Bar plots indicate the mean of three biological replicates VSports. Error bars indicate the standard deviation. .
Setdb1 mediates SHIN-induced H3K9me3. HA36::SHIN cells were stably transduced with a Cas9 expression vector and then transduced with sgRNAs against Setdb1 or a control sequence, respectively. ChIP-qPCR for H3K9me3 was performed 4 days after sgRNA transduction VSports app下载. Since we could not select for Setdb1 knockout, the chromatin isolate for ChIP is composed of deleted and non-deleted cells. RMCE PCR amplicons (1–3) are indicated. Control regions: maj sat, major satellite repeats; pos, Polrmt; neg, Tia1; and IAP, endogenous IAP elements. Bar plots indicate the mean of three biological replicates. Error bars indicate the standard deviation. .
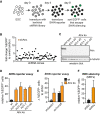
Scheme of the shRNA screen. ES cells were transduced with a genome-wide pooled shRNA library at a low MOI and then transduced with the SHIN reporter. Cells that escaped SHIN silencing were isolated by FACS sorting based on their high EGFP fluorescence. shRNA sequences of the EGFP+ cells were amplified by PCR and cloned into a lentiviral vector.
SHIN reporter assay in ES cells that were transduced with 71 random shRNA clones from the primary screen and scrambled control shRNAs. Dot plot displays the ratio of EGFP+ cells in test shRNA versus control shRNA transductions. The test shRNA with strongest reduction in SHIN silencing was directed against Atrx (yellow dot). The top-scoring shRNAs of the secondary screen are listed in Supplementary Table S2.
Western blot for Atrx in wild-type and four independent Atrx ko cell lines. Tubulin serves as a loading control.
SHIN reporter silencing depends on Atrx. Wild-type and four independent Atrx ko ES cell lines were transduced with the SHIN reporter or control vector, and EGFP fluorescence was monitored 2 days later by FACS. The percentage of EGFP-positive cells after SHIN reporter transduction is shown relative to the control transduction (relative % EGFP+ cells). The bar plot indicates the mean of three biological replicates. Error bars indicate the standard deviation.
Atrx ko cells are sensitive to perturbations in the Setdb1/Trim28 silencing pathway. SHIN reporter silencing in wild-type and Atrx ko ES cells that were transduced with control (shScr), Setdb1, and Trim28 shRNAs. Bar diagram shows the difference in EGFP-positive cells upon knockdown of Setdb1 and Trim28 compared to control-treated cells. Bar plots indicate the mean of three to six biological replicates. Error bars indicate the standard deviation.
Atrx is generally important for Setdb1-/Trim28-dependent silencing. Mouse embryonic fibroblasts were transduced with control and Atrx shRNAs. Silencing of an EGFP reporter construct containing the repressor binding site of the MLV retrovirus was monitored. The percentage of EGFP-positive cells after reporter transduction is shown relative to the control transduction (relative % EGFP+ cells). The bar plot indicates the mean of three biological replicates. Error bars indicate the standard deviation.

Scheme of the re-silencing assay in HA36::SHIN cells. The silenced transgene in HA36::SHIN cells was reactivated by the addition of doxycycline which activates the Tet-responsive promoter (TRE). Cells in which the reporter was immediately activated (approx. 2–3%) were selected with zeocin. The EGFP+ cells were then transduced with control or Atrx shRNAs. Five days later, re-silencing was triggered by the removal of the zeocin selection pressure (day 0), and EGFP expression was monitored in the presence or absence of doxycycline during the next 4 days (day 1–day 4).
Atrx is necessary for efficient silencing in competition with transcriptional activity. Re-silencing experiments as outlined in (A) were performed. Top panel: histograms displaying EGFP fluorescence during the course of re-silencing. Removal of doxycycline (−dox) inactivates the TRE promoter, resulting in loss of EGFP expression. Despite maintained TRE promoter stimulation (+dox), a large number of control cells (shScr) can silence the reporter, while Atrx-depleted cells (shAtrx) are unable to induce silencing. Lower panel: Line plots indicating the average of the median EGFP fluorescence in three to four biological replicates. Error bars indicate the standard deviation.
Atrx knockdown leads to impaired heterochromatin establishment. ChIP-qPCR analysis of H3K9me3 and H3K4me3 across the RMCE locus in HA36::SHIN cells at day 4 of the re-silencing assays as outlined in (A). Positions of the primer pairs at the RMCE locus are indicated in (A). IAP, endogenous IAP elements. H3K9me3 ChIP: pos, Polrmt; neg, Tia1; IAP, global endogenous IAPs. H3K4me3 ChIP: pos, Tia1; neg, Polrmt; IAP, global endogenous IAPs. Mock (beads only control): pos, Polrmt; neg, Tia1. Bar plots indicate the mean of three biological replicates. Error bars indicate the standard deviation.

Atrx binds to IAP retrotransposons. Cumulative ChIP-seq coverage profiles across IAP elements for Atrx, Setdb1, Trim28, and H3K9me3. The structure of IAP elements is shown schematically; the position of the SHIN sequence is marked in dark gray. rpkm, reads per kilobase per million of reads.
H3K9me3 is not altered in Atrx ko cells. ChIP-qPCR analysis for Atrx and H3K9me3 in wild-type (wt) and Atrx ko ES cells. pos, Polrmt; neg, Tia1; IAP, global endogenous IAPs; and IAP SHIN, SHIN sequence of IAP elements. Bar plots indicate the mean of three biological replicates. Error bars indicate the standard error of the mean.
Atrx is recruited to newly formed heterochromatic sites. Atrx ChIP was performed in HA36::SHIN cells, in which the SHIN reporter locus was newly integrated into a defined locus by RMCE (see Fig2B). Bar plots indicate the mean of three biological replicates. Error bars indicate the standard deviation.
Setdb1, Atrx, H3K9me3, and Trim28 co-occupy distinct ERV elements. Binary heatmap showing ChIP-seq enrichment over input (> 1.5-fold) on all mouse ERV repeat classes. Selected ERV classes are indicated. Complete information is provided in Supplementary Fig S6 and Supplementary Table S4.
MusD/ETn retrotransposons are de-repressed in Atrx ko cells. RT–qPCR analysis of selected retrotransposon classes in wild-type (wt) and Atrx ko ES cells. Bar plots indicate the mean of three to eleven biological replicates. Error bars indicate the standard deviation.
Heterochromatin maintenance is compromised in Atrx ko ES cells when core heterochromatin proteins are depleted. Wild-type and Atrx ko ES cells were transduced with control or Trim28 shRNAs, and the expression of endogenous IAP elements was measured by qRT–PCR. Bar plots indicate the mean of three biological replicates. Error bars indicate the standard deviation.
Heterochromatin is more vulnerable in Atrx ko cells. Addition of doxycycline to HA36::SHIN cells results in binding of the reverse Tet transactivator (rtTA) to the Tet-responsive promoter (TRE) in the SHIN reporter locus. Percentage of EGFP-expressing cells was monitored before and after the addition of doxycycline. Bar plots indicate the mean of three biological replicates. Error bars indicate the standard deviation.
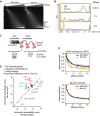
Chromatin accessibility is not globally changed in Atrx ko cells. Nuclei of wild-type and Atrx ko ES cells were digested with increasing amounts of MNase (0–16 U), and DNA was analyzed on an agarose gel stained with ethidium bromide. M, size marker; “-”, no MNase; size of mono-, di-, tri-, and tetra-nucleosomes is indicated.
Overlay of DNA electropherograms obtained from Bioanalyzer runs of the MNase-digested DNA from wild-type and Atrx ko ES cells. The first and the last sharp peak represent the markers of the Agilent DNA 12000 kit.
Schematic of the locus-specific chromatin accessibility assay. MNase cuts open chromatin faster than more compact, less accessible regions. The amount of uncut DNA of a specific locus thus correlates with accessibility of this region and can be measured by qPCR.
Heterochromatin on IAP elements is less compact in Atrx ko cells. Locus-specific chromatin accessibility assays were performed in wild-type and Atrx ko ES cells using a range of MNase concentrations. Curves represent an example of a smoothing fit to the data points. Shaded areas demarcate a confidence interval based on ± one functional standard deviation.
Generally increased heterochromatin accessibility in Atrx ko cells. Locus-specific chromatin accessibility assays were performed in wild-type and Atrx ko ES cells. Curve fitting to the data points resulted in a chromatin accessibility score which positively correlates with chromatin accessibility (see Supplementary Materials and Methods). The average chromatin accessibility score for indicated regions in wild-type and Atrx ko cells was plotted as dots from three biological replicates. The entire calculated curve fits from three biological replicates were used to assess statistically significant differences in digestion behavior (shown as dots with black border). Since the chromatin accessibility score only reflects the slope of the curve fit at 50% digestion degree, this score does not directly correlate with statistically significant differences in overall digestion behavior (also see Supplementary Materials and Methods).

References (VSports app下载)
-
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. - PubMed
-
- Wilkins AS. The enemy within: an epigenetic role of retrotransposons in cancer initiation. BioEssays. 2010;32:856–865. - PubMed
-
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. - PMC (V体育平台登录) - PubMed
Publication types
- Actions (V体育安卓版)
MeSH terms
- VSports在线直播 - Actions
- V体育官网入口 - Actions
Substances (V体育平台登录)
- Actions (V体育官网)
- Actions (VSports手机版)
LinkOut - more resources
Full Text Sources
"VSports最新版本" Other Literature Sources
"V体育官网" Research Materials

