Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation
- PMID: 25926673
- PMCID: PMC4458460
- DOI: 10.4049/jimmunol.1402154
Invariant NKT cells require autophagy to coordinate proliferation and survival signals during differentiation
"VSports" Abstract
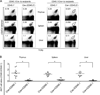
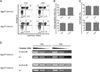
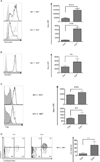
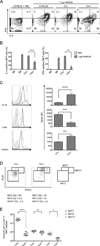
Autophagy regulates cell differentiation, proliferation, and survival in multiple cell types, including cells of the immune system. In this study, we examined the effects of a disruption of autophagy on the differentiation of invariant NKT (iNKT) cells. Using mice with a T lymphocyte-specific deletion of Atg5 or Atg7, two members of the macroautophagic pathway, we observed a profound decrease in the iNKT cell population. The deficit is cell-autonomous, and it acts predominantly to reduce the number of mature cells, as well as the function of peripheral iNKT cells. In the absence of autophagy, there is reduced progression of iNKT cells in the thymus through the cell cycle, as well as increased apoptosis of these cells. Importantly, the reduction in Th1-biased iNKT cells is most pronounced, leading to a selective reduction in iNKT cell-derived IFN-γ. Our findings highlight the unique metabolic and genetic requirements for the differentiation of iNKT cells. VSports手机版.
Copyright © 2015 by The American Association of Immunologists, Inc. V体育安卓版.
Figures





References
-
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. - PubMed (V体育官网入口)
-
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annual review of biochemistry. 2011;80:125–156. - PubMed
-
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. - "VSports" PubMed
-
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. - PMC (V体育ios版) - PubMed
Publication types
- VSports app下载 - Actions
MeSH terms (V体育官网)
- VSports - Actions
- "V体育安卓版" Actions
- Actions (V体育官网)
- Actions (V体育官网)
- V体育官网 - Actions
- Actions (VSports最新版本)
- Actions (V体育安卓版)
- Actions (V体育平台登录)
- V体育平台登录 - Actions
- "V体育安卓版" Actions
- Actions (VSports在线直播)
- "VSports手机版" Actions
- Actions (V体育平台登录)
- "V体育安卓版" Actions
- "VSports手机版" Actions
- "VSports app下载" Actions
- V体育ios版 - Actions
- VSports app下载 - Actions
Substances
- V体育官网 - Actions
- "VSports最新版本" Actions
- VSports最新版本 - Actions
- Actions (VSports app下载)
- Actions (VSports app下载)
Grants and funding
LinkOut - more resources
"VSports" Full Text Sources
Other Literature Sources

