Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge
- PMID: 25884472
- PMCID: PMC4339002
- DOI: 10.1186/s13046-015-0136-7
Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge
VSports在线直播 - Abstract
Background: Long noncoding RNAs (lncRNAs) have been identified as having functional roles in cancer biology and are deregulated in many cancers VSports手机版. The present study aimed to determine the expression, roles and functional mechanisms of a long noncoding RNA CCAT1 in the progression of hepatocellular carcinoma (HCC). .
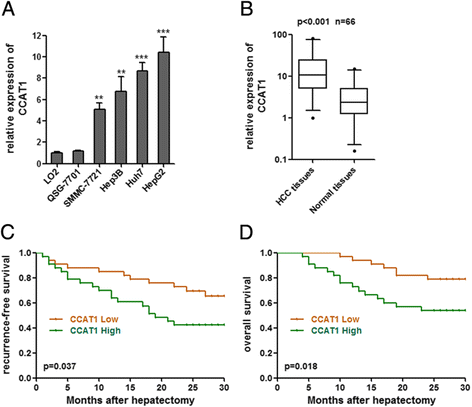
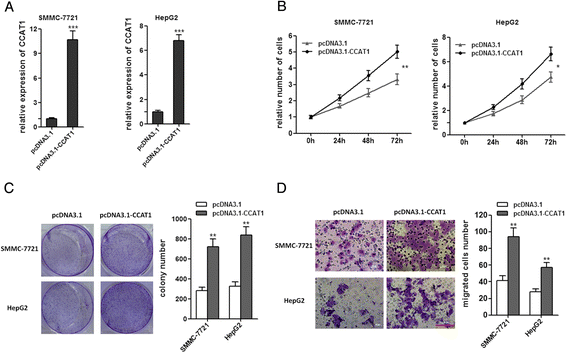
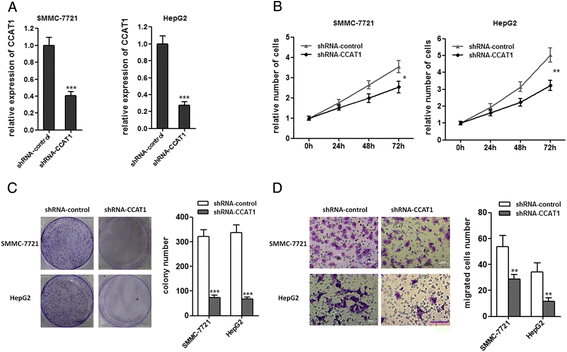
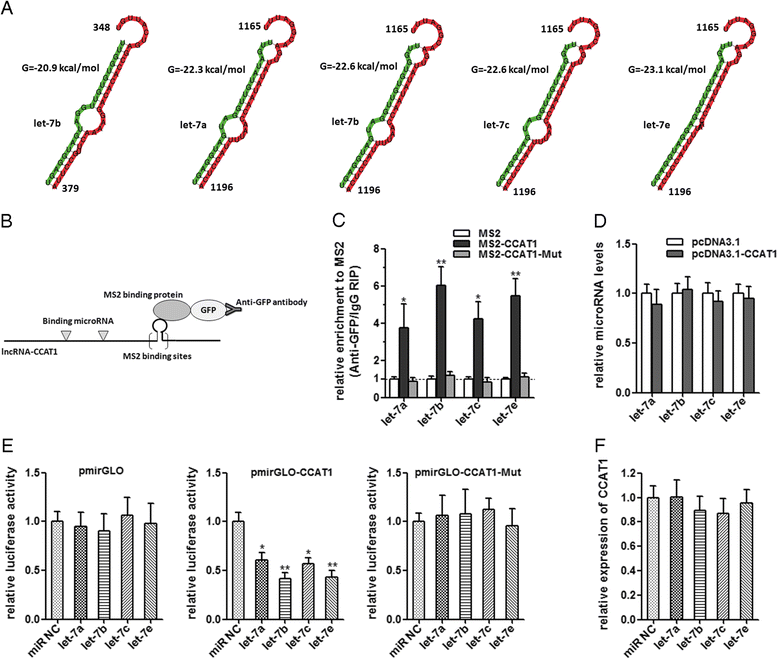
Methods: CCAT1 expression levels in 66 pairs of HCC tissues and pair-matched noncancerous hepatic tissues were tested by real-time PCR. The effects of CCAT1 on HCC cells proliferation and migration were assessed using in vitro cell proliferation and migration assays. A computational screen of microRNAs (miRNAs) target sites in CCAT1 was conducted to search for specific miRNAs binding to CCAT1. The specific binding between CCAT1 and miRNAs was confirmed by RNA immunoprecipitation assay combined with luciferase reporter assay V体育安卓版. .
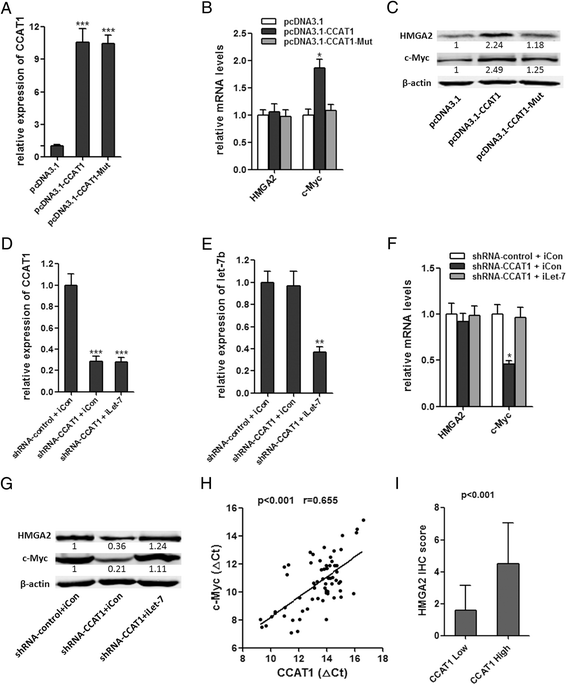
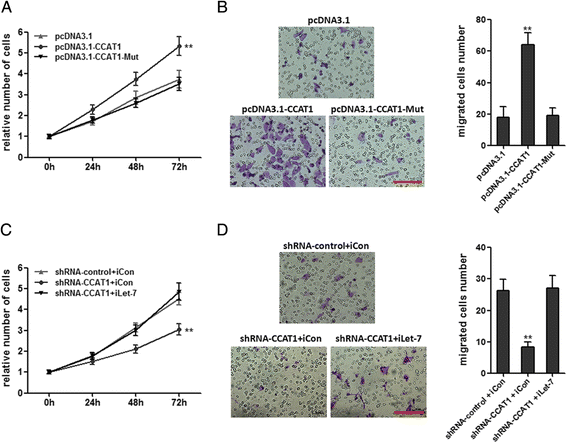
Results: CCAT1 levels are markedly increased in HCC tissues compared with pair-matched noncancerous hepatic tissues V体育ios版. Up-regulation of CCAT1 is correlated with tumor size, microvascular invasion, AFP and poor prognosis. CCAT1 promotes the proliferation and migration of HCC cells. CCAT1 functions as a molecular sponge for let-7, antagonizes its functions, and leads to the de-repression of its endogenous targets HMGA2 and c-Myc. The effect of CCAT1 on HCC cell proliferation and migration is dependent upon its competitively binding to let-7. .
Conclusions: These data suggest that CCAT1 plays a pivotal role in HCC progression via functioning as let-7 sponge, and implicate the potential application of CCAT1 for the prognosis and treatment of HCC VSports最新版本. .
"V体育官网" Figures
References
MeSH terms
- "V体育官网" Actions
- "VSports手机版" Actions
- "VSports手机版" Actions
- "VSports最新版本" Actions
- "V体育ios版" Actions
- Actions (V体育官网入口)
- "VSports注册入口" Actions
- V体育ios版 - Actions
- Actions (V体育官网)
- V体育平台登录 - Actions
- "V体育官网入口" Actions
- "V体育安卓版" Actions
Substances
- V体育平台登录 - Actions
- "V体育平台登录" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

