Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency
- PMID: 25882047
- PMCID: "VSports" PMC4816104
- DOI: 10.1038/cdd.2015.43
"V体育ios版" Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency
Abstract
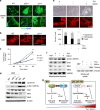
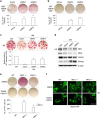
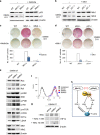
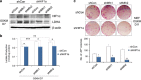
Cell reprogramming technology has allowed the in vitro control of cell fate transition, thus allowing for the generation of highly desired cell types to recapitulate in vivo developmental processes and architectures. However, the precise molecular mechanisms underlying the reprogramming process remain to be defined. Here, we show that depleting p53 and p21, which are barriers to reprogramming, yields a high reprogramming efficiency. Deletion of these factors results in a distinct mitochondrial background with low expression of oxidative phosphorylation subunits and mitochondrial fusion proteins, including mitofusin 1 and 2 (Mfn1/2). Importantly, Mfn1/2 depletion reciprocally inhibits the p53-p21 pathway and promotes both the conversion of somatic cells to a pluripotent state and the maintenance of pluripotency. Mfn1/2 depletion facilitates the glycolytic metabolic transition through the activation of the Ras-Raf and hypoxia-inducible factor 1α (HIF1α) signaling at an early stage of reprogramming. HIF1α is required for increased glycolysis and reprogramming by Mfn1/2 depletion VSports手机版. Taken together, these results demonstrate that Mfn1/2 constitutes a new barrier to reprogramming, and that Mfn1/2 ablation facilitates the induction of pluripotency through the restructuring of mitochondrial dynamics and bioenergetics. .
VSports最新版本 - Figures




References
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. - PubMed
-
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009; 460: 1140–1144. - V体育平台登录 - PMC - PubMed
"V体育2025版" Publication types
- VSports app下载 - Actions
MeSH terms
- Actions (V体育安卓版)
- Actions (V体育官网入口)
- V体育2025版 - Actions
- "VSports手机版" Actions
- "VSports手机版" Actions
- VSports手机版 - Actions
- V体育官网入口 - Actions
- VSports在线直播 - Actions
- "V体育安卓版" Actions
V体育官网 - Substances
- "VSports在线直播" Actions
- Actions (VSports注册入口)
- VSports - Actions
- "V体育官网" Actions
- Actions (V体育官网)
- "VSports注册入口" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
VSports手机版 - Molecular Biology Databases
Research Materials
Miscellaneous

