A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin
- PMID: 25778913
- PMCID: PMC4448683
- DOI: 10.1101/gr.188870.114
A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin
"V体育2025版" Abstract
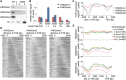
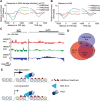
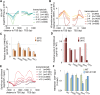
Nucleosome composition actively contributes to chromatin structure and accessibility. Cells have developed mechanisms to remove or recycle histones, generating a landscape of differentially aged nucleosomes. This study aimed to create a high-resolution, genome-wide map of nucleosome turnover in Schizosaccharomyces pombe. The recombination-induced tag exchange (RITE) method was used to study replication-independent nucleosome turnover through the appearance of new histone H3 and the disappearance or preservation of old histone H3. The genome-wide location of histones was determined by chromatin immunoprecipitation-exonuclease methodology (ChIP-exo). The findings were compared with diverse chromatin marks, including histone variant H2A. Z, post-translational histone modifications, and Pol II binding. Finally, genome-wide mapping of the methylation states of H4K20 was performed to determine the relationship between methylation (mono, di, and tri) of this residue and nucleosome turnover. Our analysis showed that histone recycling resulted in low nucleosome turnover in the coding regions of active genes, stably expressed at intermediate levels. High levels of transcription resulted in the incorporation of new histones primarily at the end of transcribed units VSports手机版. H4K20 was methylated in low-turnover nucleosomes in euchromatic regions, notably in the coding regions of long genes that were expressed at low levels. This transcription-dependent accumulation of histone methylation was dependent on the histone chaperone complex FACT. Our data showed that nucleosome turnover is highly dynamic in the genome and that several mechanisms are at play to either maintain or suppress stability. In particular, we found that FACT-associated transcription conserves histones by recycling them and is required for progressive H4K20 methylation. .
© 2015 Svensson et al. ; Published by Cold Spring Harbor Laboratory Press V体育安卓版. .
Figures



References (V体育官网入口)
-
- Aygün O, Mehta S, Grewal SI. 2013. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol 20: 547–554. - "VSports app下载" PMC - PubMed
-
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093. - PubMed
Publication types
MeSH terms
- Actions (VSports注册入口)
- "VSports" Actions
- "V体育ios版" Actions
- Actions (VSports注册入口)
- Actions (V体育平台登录)
- "VSports最新版本" Actions
- Actions (V体育官网入口)
- Actions (VSports)
- Actions (VSports手机版)
- "VSports注册入口" Actions
- "VSports手机版" Actions
Substances
- "VSports手机版" Actions
V体育平台登录 - Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
