Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells
- PMID: 25760330
- PMCID: "V体育ios版" PMC4356518
- DOI: VSports - 10.1371/journal.ppat.1004712
"VSports在线直播" Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells
V体育安卓版 - Abstract
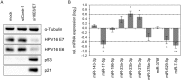
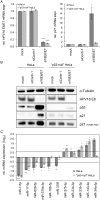
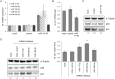
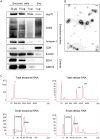
Specific types of human papillomaviruses (HPVs) cause cervical cancer. Cervical cancers exhibit aberrant cellular microRNA (miRNA) expression patterns. By genome-wide analyses, we investigate whether the intracellular and exosomal miRNA compositions of HPV-positive cancer cells are dependent on endogenous E6/E7 oncogene expression. Deep sequencing studies combined with qRT-PCR analyses show that E6/E7 silencing significantly affects ten of the 52 most abundant intracellular miRNAs in HPV18-positive HeLa cells, downregulating miR-17-5p, miR-186-5p, miR-378a-3p, miR-378f, miR-629-5p and miR-7-5p, and upregulating miR-143-3p, miR-23a-3p, miR-23b-3p and miR-27b-3p. The effects of E6/E7 silencing on miRNA levels are mainly not dependent on p53 and similarly observed in HPV16-positive SiHa cells. The E6/E7-regulated miRNAs are enriched for species involved in the control of cell proliferation, senescence and apoptosis, suggesting that they contribute to the growth of HPV-positive cancer cells. Consistently, we show that sustained E6/E7 expression is required to maintain the intracellular levels of members of the miR-17~92 cluster, which reduce expression of the anti-proliferative p21 gene in HPV-positive cancer cells. In exosomes secreted by HeLa cells, a distinct seven-miRNA-signature was identified among the most abundant miRNAs, with significant downregulation of let-7d-5p, miR-20a-5p, miR-378a-3p, miR-423-3p, miR-7-5p, miR-92a-3p and upregulation of miR-21-5p, upon E6/E7 silencing. Several of the E6/E7-dependent exosomal miRNAs have also been linked to the control of cell proliferation and apoptosis. This study represents the first global analysis of intracellular and exosomal miRNAs and shows that viral oncogene expression affects the abundance of multiple miRNAs likely contributing to the E6/E7-dependent growth of HPV-positive cancer cells. VSports手机版.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- zur Hausen H (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2: 342–350. - PubMed
-
- von Knebel Doeberitz M, Oltersdorf T, Schwarz E, Gissmann L (1988) Correlation of modified human papilloma virus early gene expression with altered growth properties in C4–1 cervical carcinoma cells. Cancer Res 48: 3780–3786. - "VSports在线直播" PubMed
-
- McLaughlin-Drubin ME, Munger K (2009) Oncogenic activities of human papillomaviruses. Virus Res 143: 195–208. 10.1016/j.virusres.2009.06.008 - "V体育ios版" DOI - PMC - PubMed
-
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63: 1129–1136. - PubMed
-
- Klingelhutz AJ, Foster SA, McDougall JK (1996) Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380: 79–82. - PubMed
Publication types
"VSports在线直播" MeSH terms
- "VSports app下载" Actions
- "V体育官网" Actions
- V体育2025版 - Actions
- V体育安卓版 - Actions
- Actions (V体育平台登录)
- Actions (VSports注册入口)
- VSports注册入口 - Actions
- "VSports在线直播" Actions
- Actions (VSports手机版)
- "V体育ios版" Actions
- "V体育安卓版" Actions
- "VSports最新版本" Actions
Substances
- "V体育官网" Actions
- Actions (VSports注册入口)
LinkOut - more resources
Full Text Sources
V体育ios版 - Other Literature Sources
"VSports最新版本" Medical
Molecular Biology Databases (V体育2025版)
Research Materials
Miscellaneous

