miR-503 represses human cell proliferation and directly targets the oncogene DDHD2 by non-canonical target pairing
- PMID: 25653011
- PMCID: V体育官网入口 - PMC4326481
- DOI: 10.1186/s12864-015-1279-9
miR-503 represses human cell proliferation and directly targets the oncogene DDHD2 by non-canonical target pairing
"V体育2025版" Abstract
Background: The pathways regulating the transition of mammalian cells from quiescence to proliferation are mediated by multiple miRNAs VSports手机版. Despite significant improvements in our understanding of miRNA targeting, the majority of miRNA regulatory networks are still largely unknown and require experimental validation. .
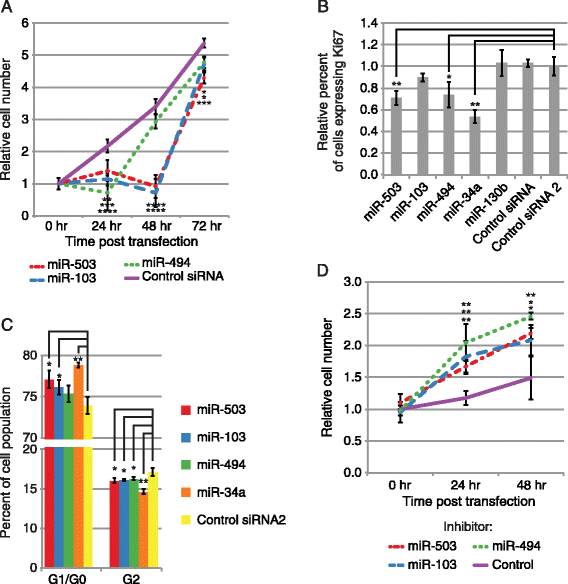
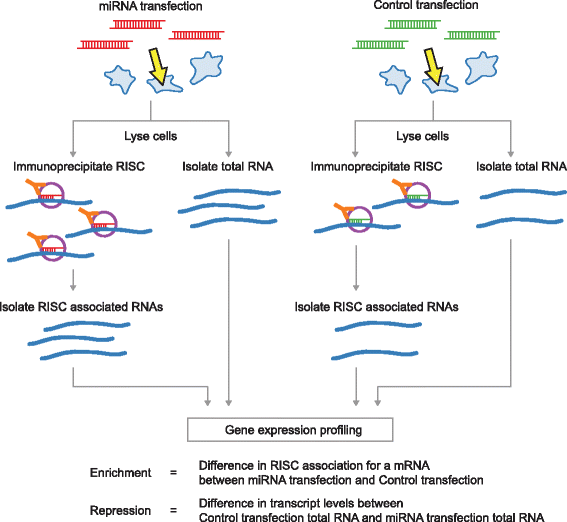
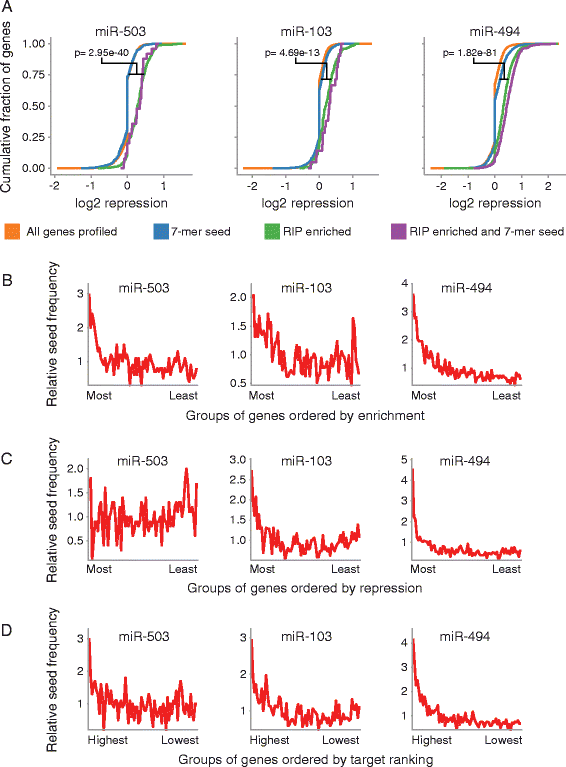
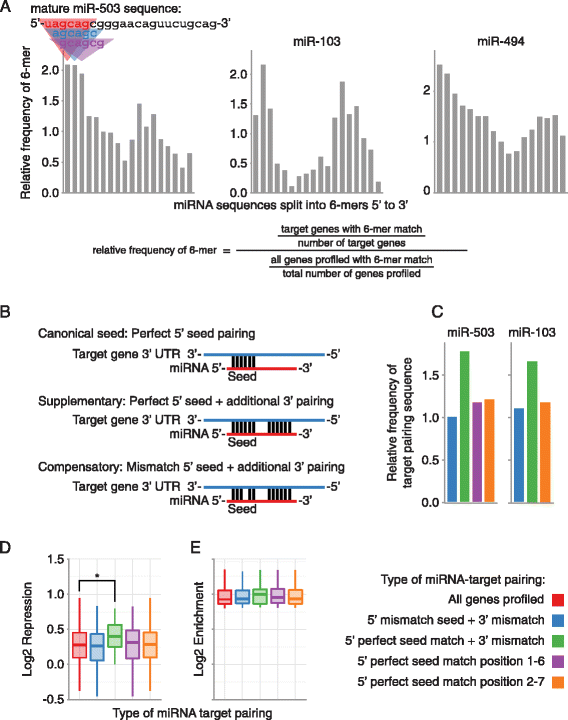
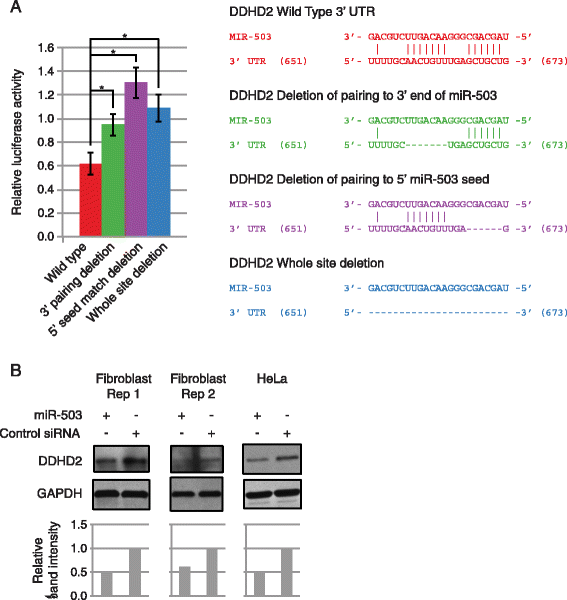
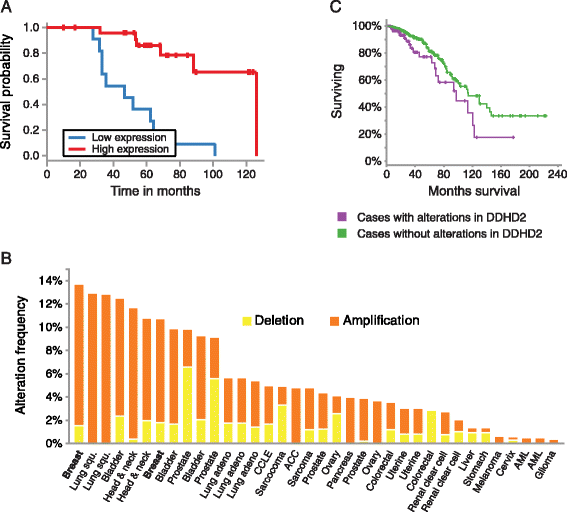
Results: Here we identified miR-503, miR-103, and miR-494 as negative regulators of proliferation in primary human cells. We experimentally determined their genome wide target profiles using RNA-induced silencing complex (RISC) immunoprecipitations and gene expression profiling V体育安卓版. Analysis of the genome wide target profiles revealed evidence of extensive regulation of gene expression through non-canonical target pairing by miR-503. We identified the proto-oncogene DDHD2 as a target of miR-503 that requires pairing outside of the canonical 5' seed region of miR-503, representing a novel mode of miRNA-target pairing. Further bioinformatics analysis implicated miR-503 and DDHD2 in breast cancer tumorigenesis. .
Conclusions: Our results provide an extensive genome wide set of targets for miR-503, miR-103, and miR-494, and suggest that miR-503 may act as a tumor suppressor in breast cancer by its direct non-canonical targeting of DDHD2. V体育ios版.
Figures
References
-
- Liu H, Adler AS, Segal E, Chang HY. A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 2007;3:e91. doi: 10.1371/journal.pgen.0030091. - "V体育ios版" DOI - PMC - PubMed
-
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. - V体育2025版 - DOI - PMC - PubMed
Publication types
- V体育ios版 - Actions
MeSH terms
- "V体育2025版" Actions
- "V体育ios版" Actions
- "VSports注册入口" Actions
- "VSports在线直播" Actions
- VSports在线直播 - Actions
- Actions (VSports)
VSports在线直播 - Substances
- Actions (V体育2025版)
- "VSports最新版本" Actions
- Actions (VSports注册入口)
Grants and funding
"V体育官网" LinkOut - more resources
Full Text Sources
VSports手机版 - Other Literature Sources
Medical
Molecular Biology Databases

