DNA is an antimicrobial component of neutrophil extracellular traps
- PMID: 25590621
- PMCID: PMC4295883
- DOI: 10.1371/journal.ppat.1004593
DNA is an antimicrobial component of neutrophil extracellular traps
"VSports" Abstract
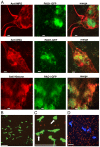
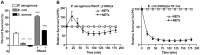
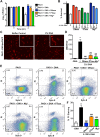
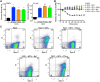
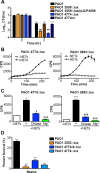
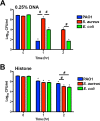
Neutrophil extracellular traps (NETs) comprise an ejected lattice of chromatin enmeshed with granular and nuclear proteins that are capable of capturing and killing microbial invaders. Although widely employed to combat infection, the antimicrobial mechanism of NETs remains enigmatic VSports手机版. Efforts to elucidate the bactericidal component of NETs have focused on the role of NET-bound proteins including histones, calprotectin and cathepsin G protease; however, exogenous and microbial derived deoxyribonuclease (DNase) remains the most potent inhibitor of NET function. DNA possesses a rapid bactericidal activity due to its ability to sequester surface bound cations, disrupt membrane integrity and lyse bacterial cells. Here we demonstrate that direct contact and the phosphodiester backbone are required for the cation chelating, antimicrobial property of DNA. By treating NETs with excess cations or phosphatase enzyme, the antimicrobial activity of NETs is neutralized, but NET structure, including the localization and function of NET-bound proteins, is maintained. Using intravital microscopy, we visualized NET-like structures in the skin of a mouse during infection with Pseudomonas aeruginosa. Relative to other bacteria, P. aeruginosa is a weak inducer of NETosis and is more resistant to NETs. During NET exposure, we demonstrate that P. aeruginosa responds by inducing the expression of surface modifications to defend against DNA-induced membrane destabilization and NET-mediated killing. Further, we show induction of this bacterial response to NETs is largely due to the bacterial detection of DNA. Therefore, we conclude that the DNA backbone contributes both to the antibacterial nature of NETs and as a signal perceived by microbes to elicit host-resistance strategies. .
Conflict of interest statement
The authors have declared that no competing interests exist.
V体育官网入口 - Figures

"VSports app下载" Comment in
-
Novel role of DNA in neutrophil extracellular traps.Trends Microbiol. 2015 Jun;23(6):330-1. doi: 10.1016/j.tim.2015.04.003. Epub 2015 Apr 23. Trends Microbiol. 2015. PMID: 25913613
V体育平台登录 - References
-
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, et al. (2004) Neutrophil extracellular traps kill bacteria. Science 303(5663): 1532–1535. 10.1126/science.1092385 - "VSports app下载" DOI - PubMed
V体育2025版 - Publication types
- "VSports最新版本" Actions
"VSports" MeSH terms
- Actions (V体育官网)
- "VSports注册入口" Actions
"V体育平台登录" Substances
"VSports最新版本" LinkOut - more resources
Full Text Sources
Other Literature Sources

