"V体育官网入口" Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice
- PMID: 25567988
- PMCID: "V体育ios版" PMC4333419
- DOI: V体育安卓版 - 10.1093/nar/gku1384
Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice (V体育2025版)
Abstract
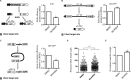
DNA double strand breaks are the most cytotoxic lesions that can occur on the DNA. They can be repaired by different mechanisms and optimal survival requires a tight control between them. Here we uncover protein deneddylation as a major controller of repair pathway choice VSports手机版. Neddylation inhibition changes the normal repair profile toward an increase on homologous recombination. Indeed, RNF111/UBE2M-mediated neddylation acts as an inhibitor of BRCA1 and CtIP-mediated DNA end resection, a key process in repair pathway choice. By controlling the length of ssDNA produced during DNA resection, protein neddylation not only affects the choice between NHEJ and homologous recombination but also controls the balance between different recombination subpathways. Thus, protein neddylation status has a great impact in the way cells respond to DNA breaks. .
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research V体育安卓版. .
Figures
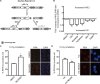
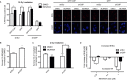
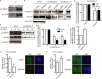
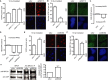
References
-
- Aguilera A., Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. - PubMed
-
- Jackson S.P., Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. - PubMed (V体育平台登录)
Publication types (VSports)
VSports - MeSH terms
- "V体育ios版" Actions
- Actions (V体育2025版)
- Actions (VSports)
- Actions (V体育官网)
- VSports - Actions
- VSports在线直播 - Actions
Substances
- Actions (VSports)
- "VSports在线直播" Actions
- V体育ios版 - Actions
LinkOut - more resources
Full Text Sources (V体育官网)
Other Literature Sources
Miscellaneous

