ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
- PMID: 25538301
- PMCID: PMC4460490
- DOI: 10.1073/pnas.1411258112
ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Erratum in
-
VSports - Correction for Noh et al., ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons.Proc Natl Acad Sci U S A. 2018 Feb 6;115(6):E1329. doi: 10.1073/pnas.1800522115. Epub 2018 Jan 30. Proc Natl Acad Sci U S A. 2018. PMID: 29382748 Free PMC article. No abstract available.
Abstract
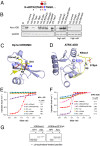
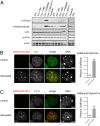
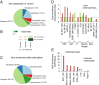
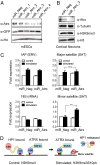
ATRX (the alpha thalassemia/mental retardation syndrome X-linked protein) is a member of the switch2/sucrose nonfermentable2 (SWI2/SNF2) family of chromatin-remodeling proteins and primarily functions at heterochromatic loci via its recognition of "repressive" histone modifications [e. g. , histone H3 lysine 9 tri-methylation (H3K9me3)]. Despite significant roles for ATRX during normal neural development, as well as its relationship to human disease, ATRX function in the central nervous system is not well understood VSports手机版. Here, we describe ATRX's ability to recognize an activity-dependent combinatorial histone modification, histone H3 lysine 9 tri-methylation/serine 10 phosphorylation (H3K9me3S10ph), in postmitotic neurons. In neurons, this "methyl/phos" switch occurs exclusively after periods of stimulation and is highly enriched at heterochromatic repeats associated with centromeres. Using a multifaceted approach, we reveal that H3K9me3S10ph-bound Atrx represses noncoding transcription of centromeric minor satellite sequences during instances of heightened activity. Our results indicate an essential interaction between ATRX and a previously uncharacterized histone modification in the central nervous system and suggest a potential role for abnormal repetitive element transcription in pathological states manifested by ATRX dysfunction. .
Keywords: ATRX; H3K9me3S10ph; crystal structure; heterochromatin; neuron V体育安卓版. .
Conflict of interest statement
The authors declare no conflict of interest.
VSports app下载 - Figures
References
-
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome) Cell. 1995;80(6):837–845. - PubMed
-
- Xie S, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236(1):87–95. - PubMed
-
- Aapola U, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65(3):293–298. - V体育安卓版 - PubMed
-
- Bérubé NG. ATRX in chromatin assembly and genome architecture during development and disease. Biochem Cell Biol. 2011;89(5):435–444. - "VSports在线直播" PubMed
Publication types
- "V体育官网" Actions
MeSH terms
- V体育ios版 - Actions
- Actions (VSports注册入口)
- Actions (VSports app下载)
- Actions (VSports手机版)
- Actions (VSports app下载)
- Actions (V体育官网)
- V体育官网 - Actions
- "VSports在线直播" Actions
- Actions (V体育平台登录)
- Actions (VSports)
- V体育2025版 - Actions
- V体育安卓版 - Actions
- "VSports" Actions
- V体育平台登录 - Actions
Substances
- "V体育2025版" Actions
- Actions (V体育平台登录)
- "VSports手机版" Actions
- "VSports" Actions
- Actions (VSports)
- V体育官网 - Actions
Associated data
- V体育官网入口 - Actions
- Actions (V体育2025版)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

