From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease
- PMID: 25426780
- PMCID: PMC4309736
- DOI: "V体育安卓版" 10.1210/er.2014-1034
From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease
Abstract
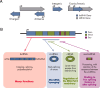
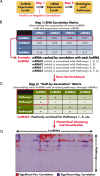
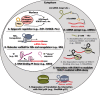
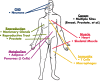
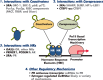
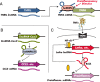
Long noncoding RNAs (lncRNAs) are a relatively poorly understood class of RNAs with little or no coding capacity transcribed from a set of incompletely annotated genes. They have received considerable attention in the past few years and are emerging as potentially important players in biological regulation. Here we discuss the evolving understanding of this new class of molecular regulators that has emerged from ongoing research, which continues to expand our databases of annotated lncRNAs and provide new insights into their physical properties, molecular mechanisms of action, and biological functions. We outline the current strategies and approaches that have been employed to identify and characterize lncRNAs, which have been instrumental in revealing their multifaceted roles ranging from cis- to trans-regulation of gene expression and from epigenetic modulation in the nucleus to posttranscriptional control in the cytoplasm. In addition, we highlight the molecular and biological functions of some of the best characterized lncRNAs in physiology and disease, especially those relevant to endocrinology, reproduction, metabolism, immunology, neurobiology, muscle biology, and cancer. Finally, we discuss the tremendous diagnostic and therapeutic potential of lncRNAs in cancer and other diseases. VSports手机版.
Figures



References
-
- Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–573. - PubMed
-
- ENCODE Project Consortium; Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. - "V体育官网入口" PMC - PubMed
-
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. - PubMed
Publication types
- "V体育官网入口" Actions
MeSH terms
- "VSports在线直播" Actions
- Actions (V体育官网)
- Actions (VSports注册入口)
- "VSports在线直播" Actions
- "VSports" Actions
- "V体育ios版" Actions
- "V体育2025版" Actions
- Actions (VSports最新版本)
- Actions (VSports app下载)
- V体育安卓版 - Actions
- Actions (V体育ios版)
Substances
- Actions (VSports最新版本)
- V体育平台登录 - Actions
- VSports最新版本 - Actions
Grants and funding
LinkOut - more resources
VSports在线直播 - Full Text Sources
Other Literature Sources
Miscellaneous

