Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma
- PMID: 25297634
- PMCID: PMC4347853
- DOI: 10.1158/0008-5472.CAN-14-1392 (V体育官网入口)
Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma (VSports app下载)
VSports在线直播 - Abstract
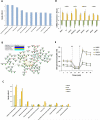
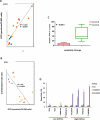
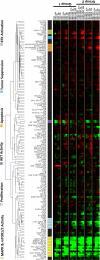
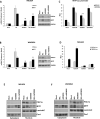
Metabolic heterogeneity is a key factor in cancer pathogenesis. We found that a subset of BRAF- and NRAS-mutant human melanomas resistant to the MEK inhibitor selumetinib displayed increased oxidative phosphorylation (OxPhos) mediated by the transcriptional coactivator PGC1α. Notably, all selumetinib-resistant cells with elevated OxPhos could be resensitized by cotreatment with the mTORC1/2 inhibitor AZD8055, whereas this combination was ineffective in resistant cell lines with low OxPhos. In both BRAF- and NRAS-mutant melanoma cells, MEK inhibition increased MITF expression, which in turn elevated levels of PGC1α. In contrast, mTORC1/2 inhibition triggered cytoplasmic localization of MITF, decreasing PGC1α expression and inhibiting OxPhos. Analysis of tumor biopsies from patients with BRAF-mutant melanoma progressing on BRAF inhibitor ± MEK inhibitor revealed that PGC1α levels were elevated in approximately half of the resistant tumors. Overall, our findings highlight the significance of OxPhos in melanoma and suggest that combined targeting of the MAPK and mTORC pathways may offer an effective therapeutic strategy to treat melanomas with this metabolic phenotype. VSports手机版.
©2014 American Association for Cancer Research. V体育安卓版.
Figures

References (V体育2025版)
-
- Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat. 2007;28:578–88. - V体育平台登录 - PubMed
-
- Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. - PubMed
-
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. - PubMed
-
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. - PMC (V体育平台登录) - PubMed
Publication types
MeSH terms
- V体育平台登录 - Actions
- "V体育安卓版" Actions
- Actions (V体育安卓版)
- VSports - Actions
- V体育官网入口 - Actions
- V体育2025版 - Actions
- Actions (V体育安卓版)
- V体育官网入口 - Actions
- Actions (VSports在线直播)
- Actions (V体育官网入口)
- V体育ios版 - Actions
Substances
- Actions (VSports注册入口)
- Actions (VSports app下载)
- "V体育安卓版" Actions
- "V体育平台登录" Actions
- Actions (V体育安卓版)
VSports - Grants and funding
LinkOut - more resources
V体育官网入口 - Full Text Sources
"V体育安卓版" Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
V体育ios版 - Miscellaneous

