Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection
- PMID: 25294838
- PMCID: PMC4231771
- DOI: 10.1093/nar/gku922
Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection
Abstract
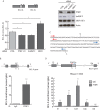
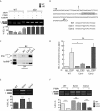
Alternative splicing (AS) modulates many physiological and pathological processes. For instance, AS of the BCL-X gene balances cell survival and apoptosis in development and cancer. Herein, we identified the polypyrimidine tract binding protein (PTBP1) as a direct regulator of BCL-X AS. Overexpression of PTBP1 promotes selection of the distal 5' splice site in BCL-X exon 2, generating the pro-apoptotic BCL-Xs splice variant. Conversely, depletion of PTBP1 enhanced splicing of the anti-apoptotic BCL-XL variant VSports手机版. In vivo cross-linking experiments and site-directed mutagenesis restricted the PTBP1 binding site to a polypyrimidine tract located between the two alternative 5' splice sites. Binding of PTBP1 to this site was required for its effect on splicing. Notably, a similar function of PTBP1 in the selection of alternative 5' splice sites was confirmed using the USP5 gene as additional model. Mechanistically, PTBP1 displaces SRSF1 binding from the proximal 5' splice site, thus repressing its selection. Our study provides a novel mechanism of alternative 5' splice site selection by PTBP1 and indicates that the presence of a PTBP1 binding site between two alternative 5' splice sites promotes selection of the distal one, while repressing the proximal site by competing for binding of a positive regulator. .
© The Author(s) 2014 V体育安卓版. Published by Oxford University Press on behalf of Nucleic Acids Research. .
Figures



References
VSports在线直播 - Publication types
MeSH terms
- Actions (V体育2025版)
- VSports手机版 - Actions
- V体育ios版 - Actions
- "VSports" Actions
- Actions (V体育官网)
- Actions (V体育平台登录)
- V体育官网入口 - Actions
- "VSports" Actions
- Actions (V体育安卓版)
Substances
- VSports最新版本 - Actions
- "VSports手机版" Actions
- V体育官网入口 - Actions
- Actions (V体育官网入口)
LinkOut - more resources
V体育官网入口 - Full Text Sources
Other Literature Sources
Molecular Biology Databases (VSports注册入口)
Research Materials
Miscellaneous

