A Bim-targeting strategy overcomes adaptive bortezomib resistance in myeloma through a novel link between autophagy and apoptosis (V体育平台登录)
- PMID: 25208888
- PMCID: "VSports" PMC4208284
- DOI: "VSports手机版" 10.1182/blood-2014-03-564534
V体育2025版 - A Bim-targeting strategy overcomes adaptive bortezomib resistance in myeloma through a novel link between autophagy and apoptosis
V体育安卓版 - Abstract
Bim contributes to resistance to various standard and novel agents. Here we demonstrate that Bim plays a functional role in bortezomib resistance in multiple myeloma (MM) cells and that targeting Bim by combining histone deacetylase inhibitors (HDACIs) with BH3 mimetics (eg, ABT-737) overcomes bortezomib resistance. BH3-only protein profiling revealed high Bim levels (Bim(hi)) in most MM cell lines and primary CD138(+) MM samples. Whereas short hairpin RNA Bim knockdown conferred bortezomib resistance in Bim(hi) cells, adaptive bortezomib-resistant cells displayed marked Bim downregulation. HDACI upregulated Bim and, when combined with ABT-737, which released Bim from Bcl-2/Bcl-xL, potently killed bortezomib-resistant cells VSports手机版. These events were correlated with Bim-associated autophagy attenuation, whereas Bim knockdown sharply increased autophagy in Bim(hi) cells. In Bim(low) cells, autophagy disruption by chloroquine (CQ) was required for HDACI/ABT-737 to induce Bim expression and lethality. CQ also further enhanced HDACI/ABT-737 lethality in bortezomib-resistant cells. Finally, HDACI failed to diminish autophagy or potentiate ABT-737-induced apoptosis in bim(-/-) mouse embryonic fibroblasts. Thus, Bim deficiency represents a novel mechanism of adaptive bortezomib resistance in MM cells, and Bim-targeting strategies combining HDACIs (which upregulate Bim) and BH3 mimetics (which unleash Bim from antiapoptotic proteins) overcomes such resistance, in part by disabling cytoprotective autophagy. .
© 2014 by The American Society of Hematology V体育安卓版. .
Figures



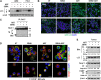
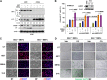


References
-
- Kumar SK, Lee JH, Lahuerta JJ, et al. International Myeloma Working Group. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group study. Leukemia. 2012;26(1):149–157. - "VSports app下载" PMC - PubMed
-
- Chauhan D, Velankar M, Brahmandam M, et al. A novel Bcl-2/Bcl-X(L)/Bcl-w inhibitor ABT-737 as therapy in multiple myeloma. Oncogene. 2007;26(16):2374–2380. - PubMed
-
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37(3):299–310. - PMC (VSports最新版本) - PubMed
Publication types
"VSports" MeSH terms
- "V体育2025版" Actions
- VSports app下载 - Actions
- V体育安卓版 - Actions
- "VSports在线直播" Actions
- "VSports最新版本" Actions
- Actions (V体育安卓版)
- Actions (V体育安卓版)
- Actions (V体育安卓版)
- VSports注册入口 - Actions
- Actions (V体育官网入口)
- "VSports" Actions
- VSports最新版本 - Actions
- "VSports最新版本" Actions
- V体育官网入口 - Actions
- "VSports最新版本" Actions
- Actions (VSports注册入口)
- VSports app下载 - Actions
- Actions (VSports注册入口)
- "V体育官网" Actions
- "V体育ios版" Actions
- Actions (VSports注册入口)
- Actions (VSports手机版)
"VSports" Substances
- V体育2025版 - Actions
- "V体育ios版" Actions
- "VSports注册入口" Actions
- Actions (VSports)
- Actions (V体育官网)
- "VSports最新版本" Actions
- Actions (VSports最新版本)
Grants and funding (V体育官网)
- P50 CA142509-01/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- P50 CA142509/CA/NCI NIH HHS/United States
- P30 NS047463/NS/NINDS NIH HHS/United States
- V体育安卓版 - R01 CA100866/CA/NCI NIH HHS/United States
- R01 CA167708/CA/NCI NIH HHS/United States
- CA100866/CA/NCI NIH HHS/United States
- P50 CA130805/CA/NCI NIH HHS/United States
- P30 CA016059/CA/NCI NIH HHS/United States
- 5P30CA016059-29/CA/NCI NIH HHS/United States
- V体育2025版 - R01 CA093738/CA/NCI NIH HHS/United States
- "V体育官网" CA167708/CA/NCI NIH HHS/United States
- "VSports最新版本" 5P30NS047463/NS/NINDS NIH HHS/United States
- CA93738/CA/NCI NIH HHS/United States
VSports在线直播 - LinkOut - more resources
Full Text Sources (VSports)
Other Literature Sources
Medical
Research Materials

