Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes
- PMID: 25159147
- PMCID: PMC4216110
- DOI: "VSports在线直播" 10.1016/j.celrep.2014.07.045
"V体育2025版" Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes
Abstract
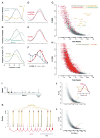
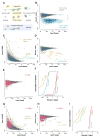
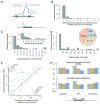
Ribosome profiling suggests that ribosomes occupy many regions of the transcriptome thought to be noncoding, including 5' UTRs and long noncoding RNAs (lncRNAs). Apparent ribosome footprints outside of protein-coding regions raise the possibility of artifacts unrelated to translation, particularly when they occupy multiple, overlapping open reading frames (ORFs). Here, we show hallmarks of translation in these footprints: copurification with the large ribosomal subunit, response to drugs targeting elongation, trinucleotide periodicity, and initiation at early AUGs. We develop a metric for distinguishing between 80S footprints and nonribosomal sources using footprint size distributions, which validates the vast majority of footprints outside of coding regions. We present evidence for polypeptide production beyond annotated genes, including the induction of immune responses following human cytomegalovirus (HCMV) infection. Translation is pervasive on cytosolic transcripts outside of conserved reading frames, and direct detection of this expanded universe of translated products enables efforts at understanding how cells manage and exploit its consequences. VSports手机版.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved V体育安卓版. .
Figures (VSports手机版)



References
-
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. - V体育安卓版 - PubMed
-
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. - PubMed
-
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. - "V体育官网入口" PMC - PubMed
Publication types
- VSports手机版 - Actions
MeSH terms
- "VSports app下载" Actions
- VSports app下载 - Actions
- "V体育平台登录" Actions
- VSports - Actions
- "V体育官网" Actions
- Actions (V体育官网)
- Actions (VSports)
- Actions (V体育官网)
- "VSports app下载" Actions
- "V体育安卓版" Actions
Substances
- Actions (V体育官网)
VSports - Associated data
Grants and funding
LinkOut - more resources
Full Text Sources (V体育官网)
Other Literature Sources
Molecular Biology Databases (V体育ios版)
Research Materials

