Bacterial phylogeny structures soil resistomes across habitats
- PMID: 24847883
- PMCID: PMC4079543
- DOI: 10.1038/nature13377
Bacterial phylogeny structures soil resistomes across habitats
Abstract
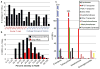
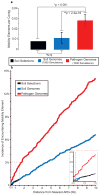
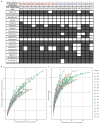
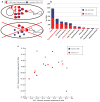
Ancient and diverse antibiotic resistance genes (ARGs) have previously been identified from soil, including genes identical to those in human pathogens. Despite the apparent overlap between soil and clinical resistomes, factors influencing ARG composition in soil and their movement between genomes and habitats remain largely unknown. General metagenome functions often correlate with the underlying structure of bacterial communities. However, ARGs are proposed to be highly mobile, prompting speculation that resistomes may not correlate with phylogenetic signatures or ecological divisions. To investigate these relationships, we performed functional metagenomic selections for resistance to 18 antibiotics from 18 agricultural and grassland soils VSports手机版. The 2,895 ARGs we discovered were mostly new, and represent all major resistance mechanisms. We demonstrate that distinct soil types harbour distinct resistomes, and that the addition of nitrogen fertilizer strongly influenced soil ARG content. Resistome composition also correlated with microbial phylogenetic and taxonomic structure, both across and within soil types. Consistent with this strong correlation, mobility elements (genes responsible for horizontal gene transfer between bacteria such as transposases and integrases) syntenic with ARGs were rare in soil by comparison with sequenced pathogens, suggesting that ARGs may not transfer between soil bacteria as readily as is observed between human pathogens. Together, our results indicate that bacterial community composition is the primary determinant of soil ARG content, challenging previous hypotheses that horizontal gene transfer effectively decouples resistomes from phylogeny. .
VSports在线直播 - Conflict of interest statement
The authors declare no competing financial interests.
VSports app下载 - Figures


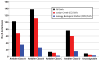
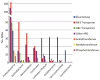
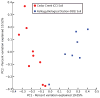
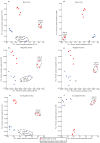
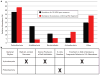


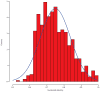
Comment in
-
Microbiology: Barriers to the spread of resistance.Nature. 2014 May 29;509(7502):567-8. doi: 10.1038/nature13342. Epub 2014 May 21. Nature. 2014. PMID: 24847882 No abstract available.
References
-
- D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. - PubMed
-
- Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. The ISME journal. 2009;3:243–251. - PubMed
-
- Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nature reviews. Microbiology. 2010;8:251–259. - VSports在线直播 - PubMed
-
- Wright GD. Antibiotic resistance in the environment: a link to the clinic? Current opinion in microbiology. 2010;13:589–594. - PubMed
Publication types
- "V体育ios版" Actions
MeSH terms
- Actions (V体育官网入口)
- Actions (VSports注册入口)
- "V体育平台登录" Actions
- "VSports最新版本" Actions
- "V体育安卓版" Actions
- VSports app下载 - Actions
- "VSports app下载" Actions
- Actions (VSports手机版)
- "VSports在线直播" Actions
- "V体育安卓版" Actions
- VSports注册入口 - Actions
- Actions (V体育平台登录)
- V体育官网入口 - Actions
- "V体育官网入口" Actions
- "V体育平台登录" Actions
V体育平台登录 - Substances
- V体育平台登录 - Actions
- VSports - Actions
- "VSports手机版" Actions
- Actions (VSports在线直播)
Associated data
- "VSports手机版" Actions
- "V体育官网" Actions
- Actions (VSports)
- Actions
- Actions
- Actions (V体育ios版)
- "V体育安卓版" Actions
- Actions
- "V体育平台登录" Actions
- "VSports在线直播" Actions
- Actions (VSports最新版本)
- V体育官网入口 - Actions
- Actions
- Actions
- Actions
- Actions
- Actions (VSports手机版)
- Actions
- Actions
- "VSports注册入口" Actions
- Actions
- Actions
- V体育安卓版 - Actions
- Actions (V体育官网入口)
- Actions (VSports最新版本)
- Actions
- "VSports在线直播" Actions
- "V体育平台登录" Actions
- VSports最新版本 - Actions
- V体育平台登录 - Actions
- Actions (VSports在线直播)
- Actions
- Actions (V体育官网)
- Actions
- "V体育官网" Actions
- Actions
- Actions
- Actions
- Actions (V体育平台登录)
- Actions
- Actions (VSports在线直播)
- "VSports手机版" Actions
- "V体育安卓版" Actions
- VSports app下载 - Actions
- Actions
- Actions (V体育安卓版)
- Actions
- "V体育平台登录" Actions
- VSports在线直播 - Actions
- "VSports在线直播" Actions
- VSports在线直播 - Actions
- "V体育安卓版" Actions
- Actions (VSports)
- Actions
- Actions
- Actions
- V体育安卓版 - Actions
- "VSports最新版本" Actions
- "VSports在线直播" Actions
- Actions
- Actions
- "V体育2025版" Actions
- Actions
- Actions
- Actions (VSports在线直播)
- Actions
- VSports - Actions
- Actions
- "VSports在线直播" Actions
- V体育平台登录 - Actions
- Actions
- "V体育官网" Actions
- VSports在线直播 - Actions
- V体育官网入口 - Actions
- "VSports手机版" Actions
- VSports - Actions
- Actions (V体育ios版)
- Actions
- Actions
- Actions (VSports app下载)
- Actions (V体育ios版)
- Actions (V体育官网入口)
- Actions
- Actions
- "VSports最新版本" Actions
- "VSports app下载" Actions
- Actions
- Actions (VSports最新版本)
- Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
- Actions (V体育官网)
- V体育官网入口 - Actions
- "V体育平台登录" Actions
- Actions (V体育平台登录)
- Actions
- Actions
- Actions (VSports手机版)
- Actions (V体育安卓版)
- Actions (VSports注册入口)
- Actions
- Actions
- Actions
- Actions (V体育官网)
- Actions (VSports)
- V体育官网入口 - Actions
- "VSports注册入口" Actions
- Actions (VSports)
- Actions (VSports)
- Actions
- VSports app下载 - Actions
- "VSports在线直播" Actions
- Actions
- V体育安卓版 - Actions
- Actions
- "V体育2025版" Actions
- Actions
- Actions
- Actions
- V体育安卓版 - Actions
- Actions
- Actions
- Actions
- "V体育平台登录" Actions
- Actions
- "V体育平台登录" Actions
- "VSports app下载" Actions
- Actions (VSports手机版)
- Actions
- "V体育安卓版" Actions
- Actions
- Actions
- "VSports app下载" Actions
- Actions (VSports app下载)
- V体育ios版 - Actions
- Actions
- "VSports最新版本" Actions
- Actions
- "VSports app下载" Actions
- Actions
- "V体育ios版" Actions
- "V体育平台登录" Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
- Actions
- Actions (V体育ios版)
- "VSports最新版本" Actions
- Actions (V体育安卓版)
- Actions (VSports最新版本)
- Actions
- "VSports" Actions
- Actions (V体育官网入口)
- "VSports" Actions
- "V体育2025版" Actions
- Actions
- V体育2025版 - Actions
- Actions (V体育官网入口)
- Actions
- "VSports最新版本" Actions
- Actions
- "V体育官网" Actions
- "V体育ios版" Actions
- Actions
- Actions
- Actions
- "V体育平台登录" Actions
- "V体育官网" Actions
- Actions (V体育平台登录)
- "V体育ios版" Actions
- Actions
- Actions
- Actions
- "VSports手机版" Actions
- Actions (V体育官网)
- V体育官网 - Actions
- "V体育2025版" Actions
- "V体育官网入口" Actions
- Actions
- "VSports最新版本" Actions
- Actions
- Actions
- V体育2025版 - Actions
- Actions (V体育安卓版)
- Actions (VSports手机版)
- Actions (V体育官网入口)
- V体育2025版 - Actions
- "V体育ios版" Actions
- Actions (VSports注册入口)
- Actions
- Actions
- V体育官网 - Actions
- V体育2025版 - Actions
- "VSports在线直播" Actions
- V体育平台登录 - Actions
- Actions (V体育ios版)
- Actions (VSports app下载)
- V体育官网 - Actions
- Actions
- Actions (V体育2025版)
- Actions (VSports)
- Actions
- Actions (VSports在线直播)
- VSports app下载 - Actions
- Actions
- Actions
- "V体育平台登录" Actions
- Actions
- Actions
- "V体育安卓版" Actions
- Actions (VSports app下载)
- Actions
- Actions
- Actions (VSports注册入口)
- VSports - Actions
- V体育安卓版 - Actions
- Actions
- VSports - Actions
- Actions
- Actions
- "V体育官网入口" Actions
- "VSports" Actions
- "V体育平台登录" Actions
- Actions
- "V体育官网入口" Actions
- "V体育ios版" Actions
- V体育官网 - Actions
- Actions (VSports最新版本)
- VSports最新版本 - Actions
- Actions (V体育官网)
- Actions
- Actions
- V体育ios版 - Actions
- Actions
- Actions
- Actions
- "V体育2025版" Actions
- Actions (VSports注册入口)
- V体育2025版 - Actions
- Actions (V体育平台登录)
- Actions (V体育平台登录)
- Actions (VSports手机版)
- "V体育安卓版" Actions
- VSports注册入口 - Actions
- VSports最新版本 - Actions
- Actions
- Actions
- "VSports手机版" Actions
- V体育2025版 - Actions
- Actions
- Actions (V体育安卓版)
- "V体育官网入口" Actions
- "V体育官网" Actions
- V体育安卓版 - Actions
- Actions
- Actions (VSports)
- "VSports" Actions
- Actions (VSports app下载)
- Actions (V体育平台登录)
- "VSports app下载" Actions
- "VSports手机版" Actions
- Actions (V体育官网入口)
- V体育ios版 - Actions
- V体育平台登录 - Actions
- Actions (VSports手机版)
- "VSports最新版本" Actions
- VSports - Actions
- VSports app下载 - Actions
- "V体育官网" Actions
- "VSports在线直播" Actions
- "V体育官网入口" Actions
- Actions
- VSports在线直播 - Actions
- Actions
- Actions
- "VSports手机版" Actions
- Actions
- "V体育官网" Actions
- VSports注册入口 - Actions
- Actions
- "V体育安卓版" Actions
- Actions (VSports)
- VSports最新版本 - Actions
- "VSports注册入口" Actions
- "V体育官网" Actions
- Actions
- VSports在线直播 - Actions
- Actions (V体育安卓版)
- "VSports最新版本" Actions
- Actions (VSports)
- Actions (V体育安卓版)
- V体育平台登录 - Actions
- "V体育安卓版" Actions
- Actions
- "V体育官网" Actions
- Actions
- "V体育官网入口" Actions
- Actions
- "V体育官网" Actions
- "VSports最新版本" Actions
- Actions
- Actions (V体育平台登录)
- V体育ios版 - Actions
- VSports在线直播 - Actions
- Actions
- Actions
- "VSports手机版" Actions
- Actions
- Actions
- Actions
- Actions
- VSports app下载 - Actions
- "VSports在线直播" Actions
- Actions (V体育官网入口)
- VSports最新版本 - Actions
- Actions
- "V体育官网" Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions (V体育安卓版)
- Actions (VSports手机版)
- "VSports手机版" Actions
- Actions
- V体育官网 - Actions
- V体育官网入口 - Actions
- Actions (VSports注册入口)
- VSports手机版 - Actions
- VSports手机版 - Actions
- "VSports在线直播" Actions
- VSports app下载 - Actions
- Actions
- Actions
- Actions (VSports在线直播)
- "V体育官网" Actions
- "VSports注册入口" Actions
- "V体育ios版" Actions
- Actions
- Actions
- VSports注册入口 - Actions
- V体育ios版 - Actions
- Actions
- Actions (VSports app下载)
- Actions (VSports手机版)
- VSports最新版本 - Actions
- "V体育2025版" Actions
- Actions
- V体育平台登录 - Actions
- "VSports在线直播" Actions
- Actions (VSports app下载)
- Actions
- Actions
- Actions (V体育平台登录)
- "VSports注册入口" Actions
- VSports在线直播 - Actions
- Actions
- Actions (V体育官网入口)
- "V体育2025版" Actions
- Actions
- VSports手机版 - Actions
- "VSports注册入口" Actions
- "V体育ios版" Actions
- VSports手机版 - Actions
- Actions
- V体育官网 - Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- "V体育官网入口" Actions
- Actions
- "V体育官网入口" Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- Actions (VSports手机版)
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- "VSports手机版" Actions
- Actions (VSports在线直播)
- V体育ios版 - Actions
- Actions
- V体育官网入口 - Actions
- "V体育安卓版" Actions
- Actions
- Actions
- V体育官网入口 - Actions
- V体育平台登录 - Actions
- "V体育官网入口" Actions
- V体育安卓版 - Actions
- Actions
- Actions (V体育平台登录)
- Actions (VSports注册入口)
- Actions
- "VSports手机版" Actions
- Actions (VSports app下载)
- VSports注册入口 - Actions
- Actions (V体育ios版)
- Actions
- Actions (VSports在线直播)
- Actions (V体育官网入口)
- V体育官网入口 - Actions
- Actions (VSports)
- Actions
- "VSports注册入口" Actions
- V体育ios版 - Actions
- "VSports在线直播" Actions
- "VSports app下载" Actions
- "VSports手机版" Actions
- V体育官网 - Actions
- Actions
- VSports最新版本 - Actions
- "VSports注册入口" Actions
- "VSports" Actions
- Actions
- VSports在线直播 - Actions
- Actions
- "VSports在线直播" Actions
- VSports手机版 - Actions
- "V体育官网入口" Actions
- "V体育官网" Actions
- Actions (VSports app下载)
- V体育2025版 - Actions
- Actions
- "V体育官网" Actions
- "V体育安卓版" Actions
- "V体育ios版" Actions
- Actions
- VSports注册入口 - Actions
- "V体育官网" Actions
- "VSports" Actions
- Actions
- "VSports app下载" Actions
- V体育2025版 - Actions
- "V体育官网" Actions
- Actions
- Actions (V体育官网)
- Actions
- Actions (VSports注册入口)
- Actions
- Actions
- Actions (VSports)
- "V体育ios版" Actions
- Actions (V体育安卓版)
- Actions (V体育官网入口)
- VSports在线直播 - Actions
- "V体育2025版" Actions
- V体育平台登录 - Actions
- Actions (V体育2025版)
- Actions
- "V体育平台登录" Actions
- Actions
- Actions
- "V体育平台登录" Actions
- Actions
- "VSports app下载" Actions
- Actions (V体育ios版)
- Actions
- "V体育安卓版" Actions
- "VSports注册入口" Actions
- Actions
- Actions
- VSports app下载 - Actions
- "VSports注册入口" Actions
- Actions
- "VSports在线直播" Actions
- Actions
- Actions
- Actions (VSports注册入口)
- "V体育官网入口" Actions
- "VSports手机版" Actions
- V体育官网入口 - Actions
- Actions (V体育平台登录)
- Actions
- Actions
- Actions
- VSports app下载 - Actions
- Actions
- "VSports app下载" Actions
- "V体育官网" Actions
- Actions
- Actions
- V体育安卓版 - Actions
- "VSports最新版本" Actions
- Actions (V体育官网)
- "VSports app下载" Actions
- Actions
- "VSports app下载" Actions
- Actions
- Actions
- Actions
- Actions (VSports最新版本)
- Actions
- Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- Actions
- VSports注册入口 - Actions
- "VSports" Actions
- Actions (V体育官网入口)
- VSports最新版本 - Actions
- VSports注册入口 - Actions
- V体育ios版 - Actions
- V体育平台登录 - Actions
- "V体育安卓版" Actions
- Actions
- VSports app下载 - Actions
- V体育ios版 - Actions
- Actions (V体育安卓版)
- Actions
- Actions
- VSports - Actions
- "V体育官网" Actions
- Actions
- Actions
- "V体育官网" Actions
- Actions
- V体育官网 - Actions
- Actions
- VSports最新版本 - Actions
- Actions
- Actions
- VSports注册入口 - Actions
- Actions
- Actions
- Actions
- V体育2025版 - Actions
- "VSports app下载" Actions
- "VSports最新版本" Actions
- Actions
- Actions
- "VSports app下载" Actions
- Actions
- V体育安卓版 - Actions
- VSports - Actions
- Actions (V体育ios版)
- Actions (V体育平台登录)
- Actions
- Actions
- Actions
- Actions (VSports最新版本)
- Actions
- "V体育ios版" Actions
- Actions
- Actions
- Actions
- VSports - Actions
- "VSports注册入口" Actions
- Actions (VSports app下载)
- VSports注册入口 - Actions
- Actions
- Actions (V体育2025版)
- Actions (V体育官网入口)
- Actions
- Actions (VSports手机版)
- "V体育安卓版" Actions
- "V体育ios版" Actions
- Actions
- Actions (VSports在线直播)
- "VSports注册入口" Actions
- VSports - Actions
- "VSports app下载" Actions
- "VSports最新版本" Actions
- "V体育安卓版" Actions
- Actions
- Actions
- Actions
- Actions (VSports app下载)
- V体育2025版 - Actions
- Actions
- Actions (VSports注册入口)
- VSports在线直播 - Actions
- Actions
- V体育平台登录 - Actions
- Actions (VSports在线直播)
- "VSports app下载" Actions
- Actions
- Actions
- V体育官网入口 - Actions
- V体育ios版 - Actions
- "VSports在线直播" Actions
- Actions (VSports app下载)
- Actions
- "VSports" Actions
- Actions
- Actions
- Actions (VSports在线直播)
- V体育2025版 - Actions
- Actions (VSports手机版)
- "V体育ios版" Actions
- "V体育2025版" Actions
- "V体育安卓版" Actions
- Actions
- Actions (V体育平台登录)
- "VSports手机版" Actions
- Actions (V体育官网)
- Actions (V体育安卓版)
- "V体育ios版" Actions
- Actions (V体育官网)
- Actions
- Actions
- V体育2025版 - Actions
- "VSports在线直播" Actions
- Actions
- Actions
- VSports app下载 - Actions
- VSports app下载 - Actions
- "V体育官网入口" Actions
- Actions
- Actions (V体育2025版)
- V体育平台登录 - Actions
- Actions (VSports在线直播)
- Actions
- Actions (V体育ios版)
- Actions
- Actions (VSports最新版本)
- Actions
- "V体育2025版" Actions
- V体育ios版 - Actions
- Actions (V体育官网入口)
- Actions
- Actions
- Actions
- VSports手机版 - Actions
- Actions (V体育安卓版)
- Actions
- Actions (VSports注册入口)
- "VSports最新版本" Actions
- Actions
- Actions
- Actions
- Actions (V体育安卓版)
- Actions (V体育官网)
- "V体育平台登录" Actions
- Actions (V体育官网)
- "V体育2025版" Actions
- "VSports app下载" Actions
- "V体育平台登录" Actions
- Actions
- Actions (VSports手机版)
- Actions
- "VSports在线直播" Actions
- V体育官网 - Actions
- "VSports注册入口" Actions
- "VSports最新版本" Actions
- Actions
- Actions (V体育平台登录)
- "VSports最新版本" Actions
- "V体育官网" Actions
- Actions
- Actions (V体育ios版)
- "V体育官网" Actions
- Actions (VSports注册入口)
- Actions (V体育官网)
- "VSports最新版本" Actions
- "VSports手机版" Actions
- "V体育平台登录" Actions
- Actions
- "VSports最新版本" Actions
- "VSports最新版本" Actions
- V体育平台登录 - Actions
- "V体育官网" Actions
- VSports手机版 - Actions
- Actions
- "VSports app下载" Actions
- VSports - Actions
- "V体育ios版" Actions
- VSports - Actions
- Actions
- Actions
- VSports注册入口 - Actions
- "V体育安卓版" Actions
- "VSports手机版" Actions
- Actions
- Actions (VSports)
- "V体育平台登录" Actions
- "V体育官网入口" Actions
- V体育官网入口 - Actions
- Actions
- "VSports手机版" Actions
- Actions
- Actions (VSports)
- Actions
- Actions
- VSports在线直播 - Actions
- Actions (V体育ios版)
- "V体育安卓版" Actions
- V体育官网 - Actions
- V体育2025版 - Actions
- Actions (VSports最新版本)
- "VSports" Actions
- VSports最新版本 - Actions
- "VSports在线直播" Actions
- Actions (VSports注册入口)
- "V体育官网" Actions
- "VSports" Actions
- Actions (VSports手机版)
- Actions
- "VSports注册入口" Actions
- Actions
- Actions (V体育平台登录)
- "V体育ios版" Actions
- "V体育官网" Actions
- Actions (V体育安卓版)
- Actions (V体育ios版)
- Actions
- "V体育安卓版" Actions
- Actions
- Actions (VSports手机版)
- Actions (VSports)
- "V体育ios版" Actions
- V体育ios版 - Actions
- Actions (V体育安卓版)
- Actions (VSports注册入口)
- "VSports在线直播" Actions
- Actions
- V体育官网 - Actions
- "V体育2025版" Actions
- "V体育安卓版" Actions
- Actions
- Actions (V体育平台登录)
- V体育2025版 - Actions
- Actions (VSports注册入口)
- VSports app下载 - Actions
- Actions
- "V体育平台登录" Actions
- Actions (VSports)
- "VSports注册入口" Actions
- Actions
- "VSports最新版本" Actions
- "VSports手机版" Actions
- Actions
- "V体育ios版" Actions
- SRA/SRP041174
"V体育平台登录" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

