MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits
- PMID: 24717937
- PMCID: PMC4011559
- DOI: 10.1038/ncomms4619
MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits
Erratum in
-
Corrigendum: MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits.Nat Commun. 2015 Oct 14;6:8679. doi: 10.1038/ncomms9679. Nat Commun. 2015. PMID: 26465271 No abstract available.
Abstract
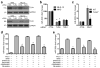
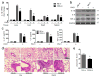
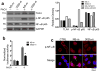
MicroRNAs (miRNAs) have been implicated in a spectrum of physiological and pathological conditions, including immune responses. miR-302b has been implicated in stem cell differentiation but its role in immunity remains unknown. Here we show that miR-302b is induced by Toll-like receptor 2 (TLR2) and TLR4 through ERK-p38-NF-κB signalling upon Gram-negative bacterium Pseudomonas aeruginosa infection. Suppression of inflammatory responses to bacterial infection is mediated by targeting IRAK4, a protein required for the activation and nuclear translocation of NF-κB. Through negative feedback, enforced expression of miR-302b or IRAK4 siRNA silencing inhibits downstream NF-κB signalling and airway leukocyte infiltration, thereby alleviating lung injury and increasing survival in P. aeruginosa-infected mice. In contrast, miR-302b inhibitors exacerbate inflammatory responses and decrease survival in P. aeruginosa-infected mice and lung cells. These findings reveal that miR-302b is a novel inflammatory regulator of NF-κB activation in respiratory bacterial infections by providing negative feedback to TLRs-mediated immunity. VSports手机版.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
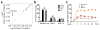




References
-
- Sun HY, Fujitani S, Quintiliani R, Yu VL. Pneumonia due to Pseudomonas aeruginosa: part II: antimicrobial resistance, pharmacodynamic concepts, and antibiotic therapy. Chest. 2011;139:1172–1185. - PubMed
Publication types
- Actions (V体育2025版)
MeSH terms
- V体育官网入口 - Actions
- VSports在线直播 - Actions
- V体育2025版 - Actions
- V体育平台登录 - Actions
- Actions (V体育官网)
- V体育官网入口 - Actions
- V体育官网 - Actions
Substances
- Actions (V体育平台登录)
- VSports手机版 - Actions
- "V体育2025版" Actions
Grants and funding
LinkOut - more resources
"V体育安卓版" Full Text Sources
Other Literature Sources
Miscellaneous

