Taxonomical and functional microbial community selection in soybean rhizosphere
- PMID: 24553468
- PMCID: PMC4817605
- DOI: "VSports app下载" 10.1038/ismej.2014.17
Taxonomical and functional microbial community selection in soybean rhizosphere
Abstract
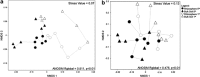
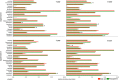
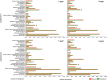
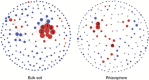
This study addressed the selection of the rhizospheric microbial community from the bulk soil reservoir under agricultural management of soybean in Amazon forest soils VSports手机版. We used a shotgun metagenomics approach to investigate the taxonomic and functional diversities of microbial communities in the bulk soil and in the rhizosphere of soybean plants and tested the validity of neutral and niche theories to explain the rhizosphere community assembly processes. Our results showed a clear selection at both taxonomic and functional levels operating in the assembly of the soybean rhizosphere community. The taxonomic analysis revealed that the rhizosphere community is a subset of the bulk soil community. Species abundance in rhizosphere fits the log-normal distribution model, which is an indicator of the occurrence of niche-based processes. In addition, the data indicate that the rhizosphere community is selected based on functional cores related to the metabolisms of nitrogen, iron, phosphorus and potassium, which are related to benefits to the plant, such as growth promotion and nutrition. The network analysis including bacterial groups and functions was less complex in rhizosphere, suggesting the specialization of some specific metabolic pathways. We conclude that the assembly of the microbial community in the rhizosphere is based on niche-based processes as a result of the selection power of the plant and other environmental factors. .
Figures


References
-
- Bastian MHS, Jacomy M. (2009). Gephi: An Open Source Software for Exploring and Manipulating Networks. International AAAI Conference on Weblogs and Social Media: San Jose, CA, USA.
-
- Benjamini Y, Hochberg Y. (1995). Controling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc 57: 289–300.
-
- Berg G, Smalla K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68: 1–13. - PubMed
-
- Brody JR, Kern SE. (2004). Sodium boric acid: Atriz-less, cooler conductive medium for DNA electrophoresis. Biotechniques 36: 214–216. - PubMed
"V体育ios版" Publication types
- V体育安卓版 - Actions
MeSH terms (VSports在线直播)
- VSports app下载 - Actions
- V体育2025版 - Actions
- "V体育平台登录" Actions
- Actions (VSports手机版)
Substances
- VSports手机版 - Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

