Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms
- PMID: 24491438
- PMCID: VSports最新版本 - PMC3992927
- DOI: 10.1158/2159-8290.CD-13-0915
Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms
Abstract
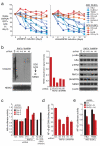
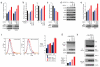
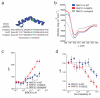
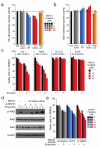
Constitutive activation of NF-κB is a hallmark of the activated B cell-like (ABC) subtype of diffuse large B-cell lymphoma (DLBCL), owing to upstream signals from the B-cell receptor (BCR) and MYD88 pathways. The linear polyubiquitin chain assembly complex (LUBAC) attaches linear polyubiquitin chains to IκB kinase-γ, a necessary event in some pathways that engage NF-κB. Two germline polymorphisms affecting the LUBAC subunit RNF31 are rare among healthy individuals (∼1%) but enriched in ABC DLBCL (7. 8%). These polymorphisms alter RNF31 α-helices that mediate binding to the LUBAC subunit RBCK1, thereby increasing RNF31-RBCK1 association, LUBAC enzymatic activity, and NF-κB engagement VSports手机版. In the BCR pathway, LUBAC associates with the CARD11-MALT1-BCL10 adapter complex and is required for ABC DLBCL viability. A stapled RNF31 α-helical peptide based on the ABC DLBCL-associated Q622L polymorphism inhibited RNF31-RBCK1 binding, decreased NF-κB activation, and killed ABC DLBCL cells, credentialing this protein-protein interface as a therapeutic target. .
Significance: We provide genetic, biochemical, and functional evidence that the LUBAC ubiquitin ligase is a therapeutic target in ABC DLBCL, the DLBCL subtype that is most refractory to current therapy. More generally, our findings highlight the role of rare germline-encoded protein variants in cancer pathogenesis. V体育安卓版.
Figures



Comment in
-
Germline polymorphisms in RNF31 regulate linear ubiquitination and oncogenic signaling.Cancer Discov. 2014 Apr;4(4):394-6. doi: 10.1158/2159-8290.CD-14-0177. Cancer Discov. 2014. PMID: 24706658
References
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. - PubMed
-
- Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappa B activity is required for survival of activated B Cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–74. - "VSports最新版本" PMC - PubMed
-
- Yang Y, Shaffer AL, 3rd, Emre NC, Ceribelli M, Zhang M, Wright G, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21:723–37. - "V体育安卓版" PMC - PubMed
-
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–10. - PubMed
Publication types
- "V体育2025版" Actions
"V体育2025版" MeSH terms
- Actions (V体育官网入口)
- "VSports app下载" Actions
- "V体育平台登录" Actions
- Actions (V体育官网)
- "V体育ios版" Actions
- "V体育ios版" Actions
- "VSports app下载" Actions
- Actions (VSports最新版本)
- VSports最新版本 - Actions
- "VSports手机版" Actions
- Actions (V体育官网)
Substances (V体育平台登录)
- Actions (V体育安卓版)
- "VSports注册入口" Actions
- V体育安卓版 - Actions
Grants and funding (V体育平台登录)
VSports在线直播 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

