Selecting age-related functional characteristics in the human gut microbiome (VSports手机版)
- PMID: 24467949
- PMCID: PMC3869192
- DOI: 10.1186/2049-2618-1-2
Selecting age-related functional characteristics in the human gut microbiome
Abstract (VSports最新版本)
Background: Human gut microbial functions are often associated with various diseases and host physiologies. Aging, a less explored factor, is also suspected to affect or be affected by microbiome alterations VSports手机版. By combining functional feature selection with supervised classification, we aim to facilitate identification of age-related functional characteristics in metagenomes from several human gut microbiome studies (MetaHIT, MicroAge, MicroObes, Kurokawa et al. 's and Gill et al. 's dataset). .
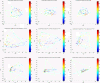
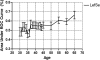
Results: We apply two feature selection methods, term frequency-inverse document frequency (TF-iDF) and minimum-redundancy maximum-relevancy (mRMR), to identify functional signatures that differentiate metagenomes by age. After features are reduced, we use a support vector machine (SVM) to predict host age of new metagenomes. Functional features are from protein families (Pfams), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, KEGG ontologies and the Gene Ontology (GO) database. Initial investigations demonstrate that ordination of the functional principal components shows great overlap between different age groups. However, when feature selection is applied, mRMR tightens the ordination cluster for each age group, and TF-iDF offers better linear separation. Both TF-iDF and mRMR were used in conjunction with a SVM classifier and achieved areas under receiver operating characteristic curves (AUCs) 10 to 15% above chance to classify individuals above/below mid-ages (about 38 to 43 years old) using Pfams. Better performance around mid-ages is also observed when using other functional categories and age-balanced dataset V体育安卓版. We also identified some age-related Pfams that improved age discrimination at age 65 with another feature selection method called LEfSe, on an age-balanced dataset. The selected functional characteristics identify a broad range of age-relevant metabolisms, such as reduced vitamin B12 synthesis, reduced activity of reductases, increased DNA damage, occurrences of stress responses and immune system compromise, and upregulated glycosyltransferases in the aging population. .
Conclusions: Feature selection can yield biologically meaningful results when used in conjunction with classification, and makes age classification of new human gut metagenomes feasible V体育ios版. While we demonstrate the promise of this approach, the data-dependent prediction performance could be further improved. We hypothesize that while the Qin et al. dataset is the most comprehensive to date, even deeper sampling is needed to better characterize and predict the microbiomes' functional content. .
V体育2025版 - Figures

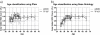
References
-
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. - "VSports最新版本" DOI - PMC - PubMed
-
- Guerrero R, Berlanga M. Life’s unity and flexibility: the ecological link. Int Microbiol. 2006;9:225–235. - PubMed
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P. et al.A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. - DOI - PMC - PubMed
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S. et al.Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. - DOI - PMC - PubMed
-
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. - DOI (V体育平台登录) - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育官网入口)
Miscellaneous

