TGF-β regulation of gene expression at early and late stages of HPV16-mediated transformation of human keratinocytes
- PMID: 24210100
- PMCID: PMC3895483
- DOI: 10.1016/j.virol.2013.08.034
"V体育平台登录" TGF-β regulation of gene expression at early and late stages of HPV16-mediated transformation of human keratinocytes
Abstract
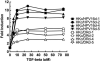
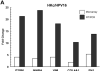
In our in vitro model for HPV16-mediated transformation, HPV16-immortalized human keratinocytes (HKc/HPV16) give rise to differentiation resistant, premalignant cells (HKc/DR). HKc/DR, but not HKc/HPV16, are resistant to growth inhibition by transforming growth factor beta (TGF-β), due to a partial loss of TGF-β receptor type I VSports手机版. We show that TGF-β activates a Smad-responsive reporter construct in HKc/DR to about 50% of the maximum levels of activation observed in HKc/HPV16. To investigate the functional significance of residual TGF-β signaling in HKc/DR, we compared gene expression profiles elicited by TGF-β treatment of HKc/HPV16 and HKc/DR on Agilent 44k human whole genome microarrays. TGF-β altered the expression of cell cycle and MAP kinase pathway genes in HKc/HPV16, but not in HKc/DR. However, epithelial-mesenchymal transition (EMT) responses to TGF-β were comparable in HKc/HPV16 and HKc/DR, indicating that the signaling pathways through which TGF-β elicits growth inhibition diverge from those that induce EMT in HPV16-transformed cells. .
Keywords: EMT; HPV; Human keratinocytes; Ski; Smad; TGF-beta V体育安卓版. .
Copyright © 2013 Elsevier Inc. All rights reserved V体育ios版. .
Figures








References (VSports注册入口)
-
- Batova A, Danielpour D, Pirisi L, Creek KE. Retinoic acid induces secretion of latent transforming growth factor beta 1 and beta 2 in normal and human papillomavirus type 16-immortalized human keratinocytes. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1992;3:763–772. - PubMed
-
- Bernat A, Avvakumov N, Mymryk JS, Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22:7871–7881. - PubMed (VSports)
-
- Bheda A, Creek KE, Pirisi L. Loss of p53 induces epidermal growth factor receptor promoter activity in normal human keratinocytes. Oncogene. 2008;27:4315–4323. - VSports手机版 - PMC - PubMed
-
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12:27–36. - VSports手机版 - PMC - PubMed
VSports在线直播 - Publication types
MeSH terms
- V体育ios版 - Actions
- "V体育官网入口" Actions
- "VSports手机版" Actions
- V体育官网入口 - Actions
- Actions (V体育ios版)
V体育官网入口 - Substances
- Actions (V体育平台登录)
"VSports app下载" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources (V体育平台登录)
"V体育平台登录" Molecular Biology Databases

