"V体育官网入口" NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies
- PMID: 24173718
- PMCID: PMC3873887 (VSports最新版本)
- DOI: 10.1091/mbc.E13-09-0558
V体育ios版 - NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies
Abstract
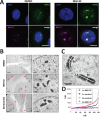
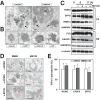
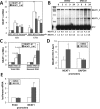
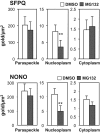
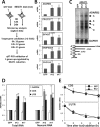
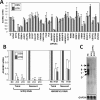
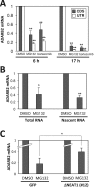
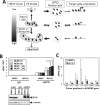
Paraspeckles are subnuclear structures formed around nuclear paraspeckle assembly transcript 1 (NEAT1)/MENε/β long noncoding RNA (lncRNA). Here we show that paraspeckles become dramatically enlarged after proteasome inhibition. This enlargement is mainly caused by NEAT1 transcriptional up-regulation rather than accumulation of undegraded paraspeckle proteins. Of interest, however, using immuno-electron microscopy, we find that key paraspeckle proteins become effectively depleted from the nucleoplasm by 50% when paraspeckle assembly is enhanced, suggesting a sequestration mechanism. We also perform microarrays from NEAT1-knockdown cells and find that NEAT1 represses transcription of several genes, including the RNA-specific adenosine deaminase B2 (ADARB2) gene. In contrast, the NEAT1-binding paraspeckle protein splicing factor proline/glutamine-rich (SFPQ) is required for ADARB2 transcription. This leads us to hypothesize that ADARB2 expression is controlled by NEAT1-dependent sequestration of SFPQ. Accordingly, we find that ADARB2 expression is strongly reduced upon enhanced SFPQ sequestration by proteasome inhibition, with concomitant reduction in SFPQ binding to the ADARB2 promoter VSports手机版. Finally, NEAT1(-/-) fibroblasts are more sensitive to proteasome inhibition, which triggers cell death, suggesting that paraspeckles/NEAT1 attenuates the cell death pathway. These data further confirm that paraspeckles are stress-responsive nuclear bodies and provide a model in which induced NEAT1 controls target gene transcription by protein sequestration into paraspeckles. .
"VSports app下载" Figures


References
-
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. - "V体育平台登录" PMC - PubMed
-
- Chen JJ, Huang WC, Chen CC. Transcriptional regulation of cyclooxygenase-2 in response to proteasome inhibitors involves reactive oxygen species-mediated signaling pathway and recruitment of CCAAT/enhancer-binding protein delta and CREB-binding protein. Mol Biol Cell. 2005;16:5579–5591. - PMC - PubMed
-
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. - PMC (V体育安卓版) - PubMed
Publication types
- Actions (V体育2025版)
MeSH terms
- VSports注册入口 - Actions
- "V体育2025版" Actions
- Actions (V体育安卓版)
- "V体育ios版" Actions
- VSports - Actions
- VSports最新版本 - Actions
- "VSports注册入口" Actions
- Actions (VSports最新版本)
- Actions (VSports手机版)
- "V体育平台登录" Actions
- "VSports注册入口" Actions
- V体育平台登录 - Actions
Substances
- Actions (VSports手机版)
- VSports最新版本 - Actions
- Actions (VSports手机版)
- VSports - Actions
- VSports app下载 - Actions
- V体育ios版 - Actions
LinkOut - more resources (VSports注册入口)
Full Text Sources
Other Literature Sources
"VSports" Molecular Biology Databases
Research Materials

