Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon (VSports注册入口)
- PMID: 23729502
- PMCID: PMC3793610
- DOI: 10.1194/jlr.M039834
Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon (VSports最新版本)
V体育安卓版 - Abstract
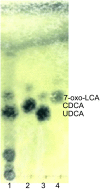
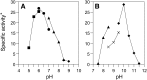
Bile acid composition in the colon is determined by bile acid flow in the intestines, the population of bile acid-converting bacteria, and the properties of the responsible bacterial enzymes. Ursodeoxycholic acid (UDCA) is regarded as a chemopreventive beneficial bile acid due to its low hydrophobicity. However, it is a minor constituent of human bile acids. Here, we characterized an UDCA-producing bacterium, N53, isolated from human feces. 16S rDNA sequence analysis identified this isolate as Ruminococcus gnavus, a novel UDCA-producer. The forward reaction that produces UDCA from 7-oxo-lithocholic acid was observed to have a growth-dependent conversion rate of 90-100% after culture in GAM broth containing 1 mM 7-oxo-lithocholic acid, while the reverse reaction was undetectable. The gene encoding 7β-hydroxysteroid dehydrogenase (7β-HSDH), which facilitates the UDCA-producing reaction, was cloned and overexpressed in Escherichia coli. Characterization of the purified 7β-HSDH revealed that the kcat/Km value was about 55-fold higher for the forward reaction than for the reverse reaction, indicating that the enzyme favors the UDCA-producing reaction. As R. gnavus is a common, core bacterium of the human gut microbiota, these results suggest that this bacterium plays a pivotal role in UDCA formation in the colon. VSports手机版.
Keywords: bile acid conversion; epimerization; intestinal bacteria; secondary bile acids V体育安卓版. .
Figures
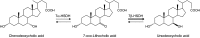




References
-
- Ridlon J. M., Kang D. J., Hylemon P. B. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259 - PubMed
-
- Begley M., Gahan C. G. M., Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29: 625–651 - PubMed
-
- Hofmann A. F. 1999. Bile acids: the good, the bad, and the ugly. News Physiol. Sci. 14: 24–29 - PubMed
-
- Bernstein H., Bernstein C., Payne C. M., Dvorakova K., Garewal H. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589: 47–65 - PubMed
"V体育官网" MeSH terms
- VSports注册入口 - Actions
- V体育官网 - Actions
- VSports - Actions
- "V体育安卓版" Actions
- "VSports app下载" Actions
- Actions (V体育平台登录)
- VSports注册入口 - Actions
- Actions (V体育ios版)
- V体育2025版 - Actions
- Actions (V体育安卓版)
Substances
- VSports在线直播 - Actions
- "V体育官网入口" Actions
- "VSports app下载" Actions
- "VSports手机版" Actions
LinkOut - more resources
Full Text Sources
"V体育官网" Other Literature Sources
"V体育安卓版" Molecular Biology Databases
Research Materials

