"VSports注册入口" Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay
- PMID: 23644405
- PMCID: PMC3781924
- DOI: 10.1053/j.gastro.2013.04.050
Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay
Abstract
Background & aims: Identification of intestinal stem cells (ISCs) has relied heavily on the use of transgenic reporters in mice, but this approach is limited by mosaic expression patterns and difficult to directly apply to human tissues VSports手机版. We sought to identify reliable surface markers of ISCs and establish a robust functional assay to characterize ISCs from mouse and human tissues. .
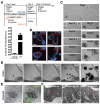
Methods: We used immunohistochemistry, real-time reverse-transcription polymerase chain reaction, and fluorescence-activated cell sorting (FACS) to analyze intestinal epithelial cells isolated from mouse and human intestinal tissues. We compared different combinations of surface markers among ISCs isolated based on expression of Lgr5-green fluorescent protein V体育安卓版. We developed a culture protocol to facilitate the identification of functional ISCs from mice and then tested the assay with human intestinal crypts and putative ISCs. .
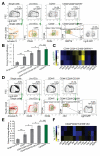
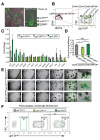
Results: CD44(+)CD24(lo)CD166(+) cells, isolated by FACS from mouse small intestine and colon, expressed high levels of stem cell-associated genes. Transit-amplifying cells and progenitor cells were then excluded based on expression of GRP78 or c-Kit. CD44(+)CD24(lo)CD166(+) GRP78(lo/-) putative stem cells from mouse small intestine included Lgr5-GFP(hi) and Lgr5-GFP(med/lo) cells V体育ios版. Incubation of these cells with the GSK inhibitor CHIR99021 and the E-cadherin stabilizer Thiazovivin resulted in colony formation by 25% to 30% of single-sorted ISCs. .
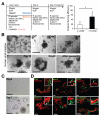
Conclusions: We developed a culture protocol to identify putative ISCs from mouse and human tissues based on cell surface markers VSports最新版本. CD44(+)CD24(lo)CD166(+), GRP78(lo/-), and c-Kit(-) facilitated identification of putative stem cells from the mouse small intestine and colon, respectively. CD44(+)CD24(-/lo)CD166(+) also identified putative human ISCs. These findings will facilitate functional studies of mouse and human ISCs. .
Keywords: CBC; CFE; CoSC; Differentiation; FACS; Flow Cytometry Analysis; GRP; IEC; IHC; ISC; PC; Paneth cell; Single-Cell Sorting; Stemness; colonic stem cell; colony-forming efficiency; crypt base columnar; fluorescence-activated cell sorting; green fluorescent protein; immunohistochemistry; intestinal epithelial cell; intestinal stem cell; qRT-PCR; quantitative reverse-transcription polymerase chain reaction. V体育平台登录.
Copyright © 2013 AGA Institute. Published by Elsevier Inc. All rights reserved. VSports注册入口.
Figures



References
-
- Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
Publication types
- "VSports注册入口" Actions
- V体育官网入口 - Actions
MeSH terms
- Actions (VSports)
- Actions (V体育平台登录)
- Actions (V体育官网)
- "VSports注册入口" Actions
- "V体育ios版" Actions
- Actions (VSports在线直播)
- VSports app下载 - Actions
- V体育平台登录 - Actions
- "VSports最新版本" Actions
Substances (V体育安卓版)
- "VSports手机版" Actions
- Actions (VSports最新版本)
- Actions (V体育平台登录)
- Actions (V体育2025版)
Grants and funding (VSports)
- U01 DK085535/DK/NIDDK NIH HHS/United States
- R01 DK091427/DK/NIDDK NIH HHS/United States
- U01DK85532/DK/NIDDK NIH HHS/United States
- U01 DK085507/DK/NIDDK NIH HHS/United States
- VSports在线直播 - U24 DK085532/DK/NIDDK NIH HHS/United States
- U01DK085507/DK/NIDDK NIH HHS/United States
- U01 DK085547/DK/NIDDK NIH HHS/United States
- U01 DK085532/DK/NIDDK NIH HHS/United States
- V体育官网 - U01DK085547/DK/NIDDK NIH HHS/United States
- "VSports最新版本" U01DK85535/DK/NIDDK NIH HHS/United States
- U01 DK085525/DK/NIDDK NIH HHS/United States
- U01DK85525/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育ios版" Research Materials
Miscellaneous

