Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins
- PMID: 23453757
- PMCID: PMC3656483
- DOI: 10.1016/j.cell.2013.02.024
Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins
Abstract
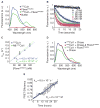
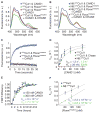
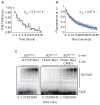
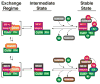
The modular SCF (Skp1, cullin, and F box) ubiquitin ligases feature a large family of F box protein substrate receptors that enable recognition of diverse targets. However, how the repertoire of SCF complexes is sustained remains unclear. Real-time measurements of formation and disassembly indicate that SCF(Fbxw7) is extraordinarily stable, but, in the Nedd8-deconjugated state, the cullin-binding protein Cand1 augments its dissociation by one-million-fold. Binding and ubiquitylation assays show that Cand1 is a protein exchange factor that accelerates the rate at which Cul1-Rbx1 equilibrates with multiple F box protein-Skp1 modules. Depletion of Cand1 from cells impedes recruitment of new F box proteins to pre-existing Cul1 and profoundly alters the cellular landscape of SCF complexes VSports手机版. We suggest that catalyzed protein exchange may be a general feature of dynamic macromolecular machines and propose a hypothesis for how substrates, Nedd8, and Cand1 collaborate to regulate the cellular repertoire of SCF complexes. .
Copyright © 2013 Elsevier Inc V体育安卓版. All rights reserved. .
Figures


Comment in
-
Cullins getting undressed by the protein exchange factor Cand1.Cell. 2013 Mar 28;153(1):14-6. doi: 10.1016/j.cell.2013.03.014. Cell. 2013. PMID: 23540687
-
Set them free: F-box protein exchange by Cand1.Cell Res. 2013 Jul;23(7):870-1. doi: 10.1038/cr.2013.55. Epub 2013 Apr 23. Cell Res. 2013. PMID: 23609796 Free PMC article.
References
-
- Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci USa. 2006;103:11515–11520. - VSports最新版本 - PMC - PubMed
-
- Bosu DR, Feng H, Min K, Kim Y, Wallenfang MR, Kipreos ET. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Developmental Biology. 2010;346:113–126. - "V体育ios版" PMC - PubMed
"VSports在线直播" Publication types
MeSH terms
- "VSports最新版本" Actions
- Actions (V体育2025版)
- "V体育ios版" Actions
- "VSports" Actions
Substances (V体育平台登录)
- "V体育安卓版" Actions
- V体育安卓版 - Actions
- "VSports在线直播" Actions
- VSports手机版 - Actions
Grants and funding
LinkOut - more resources
V体育安卓版 - Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

