"V体育安卓版" Vemurafenib potently induces endoplasmic reticulum stress-mediated apoptosis in BRAFV600E melanoma cells
- PMID: 23362240
- PMCID: "V体育官网" PMC3698985
- DOI: 10.1126/scisignal.2003057
"V体育安卓版" Vemurafenib potently induces endoplasmic reticulum stress-mediated apoptosis in BRAFV600E melanoma cells
Abstract
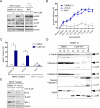
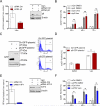
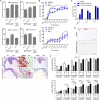
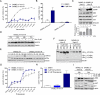
The V600E mutation in the kinase BRAF is frequently detected in melanomas and results in constitutive activation of BRAF, which then promotes cell proliferation by the mitogen-activated protein kinase signaling pathway. Although the BRAFV600E kinase inhibitor vemurafenib has remarkable antitumor activity in patients with BRAFV600E-mutated melanoma, its effects are limited by the onset of drug resistance. We found that exposure of melanoma cell lines with the BRAFV600E mutation to vemurafenib decreased the abundance of antiapoptotic proteins and induced intrinsic mitochondrial apoptosis. Vemurafenib-treated melanoma cells showed increased cytosolic concentration of calcium, a potential trigger for endoplasmic reticulum (ER) stress, which can lead to apoptosis. Consistent with an ER stress-induced response, vemurafenib decreased the abundance of the ER chaperone protein glucose-regulated protein 78, increased the abundance of the spliced isoform of the transcription factor X-box binding protein 1 (XBP1) (which transcriptionally activates genes involved in ER stress responses), increased the phosphorylation of the translation initiation factor eIF2α (which would be expected to inhibit protein synthesis), and induced the expression of ER stress-related genes. Knockdown of the ER stress response protein activating transcription factor 4 (ATF4) significantly reduced vemurafenib-induced apoptosis. Moreover, the ER stress inducer thapsigargin prevented invasive growth of tumors formed from vemurafenib-sensitive melanoma cells in vivo VSports手机版. In melanoma cells with low sensitivity or resistance to vemurafenib, combination treatment with thapsigargin augmented or induced apoptosis. Thus, thapsigargin or other inducers of ER stress may be useful in combination therapies to overcome vemurafenib resistance. .
Figures (V体育2025版)

References
-
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, Eggermont AM, Flaherty KT, Gimotty PA, Kirkwood JM, McMasters KM, Mihm MC, Jr., Morton DL, Ross MI, Sober AJ, Sondak VK. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199. published online EpubDec 20. - PMC - PubMed
-
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949. published online EpubJun 27. - PubMed
-
- Satyamoorthy K, Li G, Gerrero MR, Brose MS, Volpe P, Weber BL, Van Belle P, Elder DE, Herlyn M. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer research. 2003;63:756. published online EpubFeb 15. - PubMed
-
- Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J Clin Oncol. 2005;23:1473. published online EpubMar 1. - PubMed
-
- Meier F, Schittek B, Busch S, Garbe C, Smalley K, Satyamoorthy K, Li G, Herlyn M. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front Biosci. 2005;10:2986. - PubMed
Publication types
- "V体育官网入口" Actions
- "VSports手机版" Actions
MeSH terms (V体育安卓版)
- Actions (VSports最新版本)
- "V体育安卓版" Actions
- VSports - Actions
- Actions (VSports)
- Actions (VSports在线直播)
- VSports - Actions
- "VSports手机版" Actions
- "VSports注册入口" Actions
- "V体育官网" Actions
- V体育ios版 - Actions
- "V体育2025版" Actions
- V体育ios版 - Actions
- "V体育平台登录" Actions
"VSports注册入口" Substances
- "VSports手机版" Actions
- V体育官网 - Actions
- Actions (V体育安卓版)
- "VSports" Actions
- V体育ios版 - Actions
- "VSports" Actions
Grants and funding (V体育2025版)
LinkOut - more resources
"VSports" Full Text Sources
"V体育安卓版" Other Literature Sources
Medical
Research Materials

