Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice
- PMID: 23347395
- PMCID: PMC4053703
- DOI: 10.1186/gb-2013-14-1-r4
Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice
"VSports" Abstract
Background: Murine models are a crucial component of gut microbiome research VSports手机版. Unfortunately, a multitude of genetic backgrounds and experimental setups, together with inter-individual variation, complicates cross-study comparisons and a global understanding of the mouse microbiota landscape. Here, we investigate the variability of the healthy mouse microbiota of five common lab mouse strains using 16S rDNA pyrosequencing. .
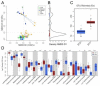
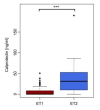
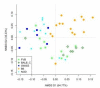
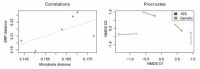
Results: We find initial evidence for richness-driven, strain-independent murine enterotypes that show a striking resemblance to those in human, and which associate with calprotectin levels, a marker for intestinal inflammation V体育安卓版. After enterotype stratification, we find that genetic, caging and inter-individual variation contribute on average 19%, 31. 7% and 45. 5%, respectively, to the variance in the murine gut microbiota composition. Genetic distance correlates positively to microbiota distance, so that genetically similar strains have more similar microbiota than genetically distant ones. Specific mouse strains are enriched for specific operational taxonomic units and taxonomic groups, while the 'cage effect' can occur across mouse strain boundaries and is mainly driven by Helicobacter infections. .
Conclusions: The detection of enterotypes suggests a common ecological cause, possibly low-grade inflammation that might drive differences among gut microbiota composition in mammals. Furthermore, the observed environmental and genetic effects have important consequences for experimental design in mouse microbiome research V体育ios版. .
"V体育安卓版" Figures
References
-
- Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One. 2008;14:e3064. doi: 10.1371/journal.pone.0003064. - VSports手机版 - DOI - PMC - PubMed
Publication types
MeSH terms
- Actions (VSports最新版本)
- "V体育ios版" Actions
- Actions (VSports注册入口)
- VSports手机版 - Actions
- V体育官网 - Actions
V体育官网入口 - Substances
LinkOut - more resources
V体育ios版 - Full Text Sources
Other Literature Sources
Miscellaneous

