Alpha thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress
- PMID: 23329831
- PMCID: PMC3585069
- DOI: 10.1074/jbc.M112.411603
"VSports注册入口" Alpha thalassemia/mental retardation syndrome X-linked gene product ATRX is required for proper replication restart and cellular resistance to replication stress
Abstract
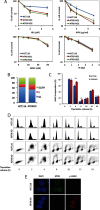
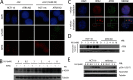
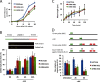
Alpha thalassemia/mental retardation syndrome X-linked (ATRX) is a member of the SWI/SNF protein family of DNA-dependent ATPases VSports手机版. It functions as a chromatin remodeler and is classified as an SNF2-like helicase. Here, we showed somatic knock-out of ATRX displayed perturbed S-phase progression as well as hypersensitivity to replication stress. ATRX is recruited to sites of DNA damage, required for efficient checkpoint activation and faithful replication restart. In addition, we identified ATRX as a binding partner of MRE11-RAD50-NBS1 (MRN) complex. Together, these results suggest a non-canonical function of ATRX in guarding genomic stability. .
Figures


V体育平台登录 - References
-
- Abraham R. T. (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196 - PubMed
-
- Zou L., Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 - PubMed
-
- Picketts D. J., Higgs D. R., Bachoo S., Blake D. J., Quarrell O. W., Gibbons R. J. (1996) ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet. 5, 1899–1907 - PubMed (V体育安卓版)
Publication types
- VSports - Actions
MeSH terms
- "VSports在线直播" Actions
- "V体育平台登录" Actions
- V体育ios版 - Actions
- "V体育安卓版" Actions
- Actions (V体育平台登录)
- "V体育平台登录" Actions
- VSports app下载 - Actions
- Actions (VSports注册入口)
- VSports - Actions
Substances
- V体育2025版 - Actions
- Actions (VSports app下载)
- Actions (V体育2025版)
- Actions (V体育平台登录)
- "VSports在线直播" Actions
Grants and funding
LinkOut - more resources
Full Text Sources (V体育官网)
Other Literature Sources
Research Materials (VSports最新版本)
Miscellaneous

