VSports在线直播 - DNA methylation profiles of long- and short-term glioblastoma survivors
- PMID: 23291739
- PMCID: PMC3592900
- DOI: "V体育ios版" 10.4161/epi.23398
VSports在线直播 - DNA methylation profiles of long- and short-term glioblastoma survivors
V体育官网入口 - Abstract
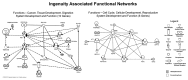
Glioblastoma (GBM) is the most common and malignant type of primary brain tumor in adults and prognosis of most GBM patients is poor VSports手机版. However, a small percentage of patients show a long term survival of 36 mo or longer after diagnosis. Epigenetic profiles can provide molecular markers for patient prognosis: recently, a G-CIMP positive phenotype associated with IDH1 mutations has been described for GBMs with good prognosis. In the present analysis we performed genome-wide DNA methylation profiling of short-term survivors (STS; overall survival < 1 y) and long-term survivors (LTS; overall survival > 3 y) by utilizing the HumanMethylation450K BeadChips to assess quantitative methylation at > 480,000 CpG sites. Cluster analysis has shown that a subset of LTS showed a G-CIMP positive phenotype that was tightly associated with IDH1 mutation status and was confirmed by analysis of the G-CIMP signature genes. Using high stringency criteria for differential hypermethylation between non-cancer brain and tumor samples, we identified 2,638 hypermethylated CpG loci (890 genes) in STS GBMs, 3,101 hypermethylated CpG loci (1,062 genes) in LTS (wild type IDH1) and 11,293 hypermethylated CpG loci in LTS (mutated for IDH1), reflecting the CIMP positive phenotype. The location of differentially hypermethylated CpG loci with respect to CpG content, neighborhood context and functional genomic distribution was similar in our sample set, with the majority of CpG loci residing in CpG islands and in gene promoters. Our preliminary study also identified a set of CpG loci differentially hypermethylated between STS and LTS cases, including members of the homeobox gene family (HOXD8, HOXD13 and HOXC4), the transcription factors NR2F2 and TFAP2A, and Dickkopf 2, a negative regulator of the wnt/β-catenin signaling pathway. .
Keywords: DNA methylation; IDH1; gliomas; long-term survivors; short-term survivors. V体育安卓版.
V体育官网 - Figures



References
-
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. - DOI (V体育平台登录) - PMC - PubMed
-
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. - V体育平台登录 - DOI - PMC - PubMed
Publication types
- "V体育官网" Actions
MeSH terms
- "VSports" Actions
- V体育ios版 - Actions
- "V体育官网" Actions
- "VSports在线直播" Actions
Substances
- "VSports注册入口" Actions
- "VSports最新版本" Actions
VSports注册入口 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials (V体育官网入口)
Miscellaneous
