"VSports注册入口" Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements
- PMID: 23201014
- PMCID: "VSports手机版" PMC3640454
- DOI: 10.1016/j.ydbio.2012.11.017
Pitx1 broadly associates with limb enhancers and is enriched on hindlimb cis-regulatory elements (V体育ios版)
"VSports最新版本" Abstract
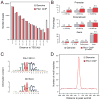
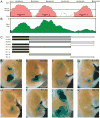
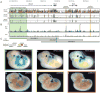
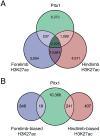
Extensive functional analyses have demonstrated that the pituitary homeodomain transcription factor Pitx1 plays a critical role in specifying hindlimb morphology in vertebrates. However, much less is known regarding the target genes and cis-regulatory elements through which Pitx1 acts. Earlier studies suggested that the hindlimb transcription factors Tbx4, HoxC10, and HoxC11 might be transcriptional targets of Pitx1, but definitive evidence for direct regulatory interactions has been lacking. Using ChIP-Seq on embryonic mouse hindlimbs, we have pinpointed the genome-wide location of Pitx1 binding sites during mouse hindlimb development and identified potential gene targets for Pitx1. We determined that Pitx1 binding is significantly enriched near genes involved in limb morphogenesis, including Tbx4, HoxC10, and HoxC11. Notably, Pitx1 is bound to the previously identified HLEA and HLEB hindlimb enhancers of the Tbx4 gene and to a newly identified Tbx2 hindlimb enhancer. Moreover, Pitx1 binding is significantly enriched on hindlimb relative to forelimb-specific cis-regulatory features that are differentially marked by H3K27ac. However, our analysis revealed that Pitx1 also strongly associates with many functionally verified limb enhancers that exhibit similar levels of activity in the embryonic mesenchyme of forelimbs and hindlimbs. We speculate that Pitx1 influences hindlimb morphology both through the activation of hindlimb-specific enhancers as well as through the hindlimb-specific modulation of enhancers that are active in both sets of limbs. VSports手机版.
Copyright © 2012 Elsevier Inc V体育安卓版. All rights reserved. .
Figures


VSports - References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. - PMC - PubMed
-
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. - "V体育官网入口" PMC - PubMed
-
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. - PMC - PubMed
-
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. - PubMed
Publication types
- Actions (V体育ios版)
- Actions (VSports app下载)
MeSH terms
- "V体育2025版" Actions
- Actions (VSports注册入口)
- "V体育安卓版" Actions
- Actions (V体育官网)
- "VSports最新版本" Actions
- VSports - Actions
- "VSports" Actions
Substances
- "VSports app下载" Actions
Grants and funding
VSports手机版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

