Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system
- PMID: 23103054
- PMCID: PMC3549668
- DOI: V体育平台登录 - 10.1016/j.stem.2012.09.011
VSports - Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system
"VSports手机版" Abstract
Although transcriptional regulation of stem cell pluripotency and differentiation has been extensively studied, only a small number of studies have addressed the roles for posttranslational modifications in these processes. A key mechanism of posttranslational modification is ubiquitination by the ubiquitin-proteasome system (UPS). Here, using shotgun proteomics, we map the ubiquitinated protein landscape during embryonic stem cell (ESC) differentiation and induced pluripotency. Moreover, using UPS-targeted RNAi screens, we identify additional regulators of pluripotency and differentiation. We focus on two of these proteins, the deubiquitinating enzyme Psmd14 and the E3 ligase Fbxw7, and characterize their importance in ESC pluripotency and cellular reprogramming. This global characterization of the UPS as a key regulator of stem cell pluripotency opens the way for future studies that focus on specific UPS enzymes or ubiquitinated substrates. VSports手机版.
Copyright © 2012 Elsevier Inc. All rights reserved V体育安卓版. .
Figures
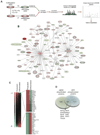
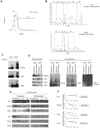
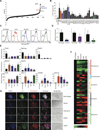

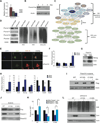

Comment in
-
UPS delivers pluripotency.Cell Stem Cell. 2012 Dec 7;11(6):728-30. doi: 10.1016/j.stem.2012.11.009. Cell Stem Cell. 2012. PMID: 23217415
References
-
- Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer investigation. 2004;22:304–311. - V体育官网入口 - PubMed
-
- An J, Yang DY, Xu QZ, Zhang SM, Huo YY, Shang ZF, Wang Y, Wu DC, Zhou PK. DNA-dependent protein kinase catalytic subunit modulates the stability of c-Myc oncoprotein. Molecular cancer. 2008;7:32. - "V体育ios版" PMC - PubMed
-
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. - PubMed
Publication types
- "VSports在线直播" Actions
MeSH terms
- "V体育平台登录" Actions
- "VSports app下载" Actions
- "V体育安卓版" Actions
- Actions (VSports手机版)
- "V体育官网" Actions
- "VSports最新版本" Actions
- Actions (V体育安卓版)
- V体育2025版 - Actions
- Actions (V体育2025版)
- Actions (V体育官网入口)
- VSports在线直播 - Actions
- "V体育官网" Actions
- V体育平台登录 - Actions
- Actions (VSports最新版本)
Substances
- Actions (V体育官网入口)
- V体育官网 - Actions
- "VSports最新版本" Actions
- VSports在线直播 - Actions
VSports app下载 - Associated data
- "VSports在线直播" Actions
Grants and funding
- "V体育ios版" 1S10 RR023680-1/RR/NCRR NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R01CA133379/CA/NCI NIH HHS/United States
- 1T32CA160002-01/CA/NCI NIH HHS/United States
- R01 CA149655/CA/NCI NIH HHS/United States
- R01 CA105129/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 5T32HL007151-33/HL/NHLBI NIH HHS/United States
- R01CA149655/CA/NCI NIH HHS/United States
- R01CA105129/CA/NCI NIH HHS/United States
- R01 GM088847/GM/NIGMS NIH HHS/United States
- P30 CA016087-30/CA/NCI NIH HHS/United States
- R01 CA133379/CA/NCI NIH HHS/United States
- V体育ios版 - T32 HL007151/HL/NHLBI NIH HHS/United States
- S10 RR023680/RR/NCRR NIH HHS/United States
- R01GM088847/GM/NIGMS NIH HHS/United States
- R01 CA155125/CA/NCI NIH HHS/United States
- T32 CA160002/CA/NCI NIH HHS/United States
- 1U19A1091175-01/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

