Species-level analysis of DNA sequence data from the NIH Human Microbiome Project
- PMID: 23071716
- PMCID: PMC3468466
- DOI: "VSports在线直播" 10.1371/journal.pone.0047075
Species-level analysis of DNA sequence data from the NIH Human Microbiome Project
Abstract
Background: Outbreaks of antibiotic-resistant bacterial infections emphasize the importance of surveillance of potentially pathogenic bacteria. Genomic sequencing of clinical microbiological specimens expands our capacity to study cultivable, fastidious and uncultivable members of the bacterial community. Herein, we compared the primary data collected by the NIH's Human Microbiome Project (HMP) with published epidemiological surveillance data of Staphylococcus aureus. VSports手机版.
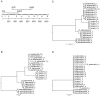
Methods: The HMP's initial dataset contained microbial survey data from five body regions (skin, nares, oral cavity, gut and vagina) of 242 healthy volunteers. A significant component of the HMP dataset was deep sequencing of the 16S ribosomal RNA gene, which contains variable regions enabling taxonomic classification. Since species-level identification is essential in clinical microbiology, we built a reference database and used phylogenetic placement followed by most recent common ancestor classification to look at the species distribution for Staphylococcus, Klebsiella and Enterococcus V体育安卓版. .
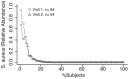
Main results: We show that selecting the accurate region of the 16S rRNA gene to sequence is analogous to carefully selecting culture conditions to distinguish closely related bacterial species. Analysis of the HMP data showed that Staphylococcus aureus was present in the nares of 36% of healthy volunteers, consistent with culture-based epidemiological data. Klebsiella pneumoniae and Enterococcus faecalis were found less frequently, but across many habitats. V体育ios版.
Conclusions: This work demonstrates that large 16S rRNA survey studies can be used to support epidemiological goals in the context of an increasing awareness that microbes flourish and compete within a larger bacterial community. This study demonstrates how genomic techniques and information could be critically important to trace microbial evolution and implement hospital infection control VSports最新版本. .
Conflict of interest statement
Figures (V体育官网入口)

References
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al.. (2011) Enterotypes of the human gut microbiome. Nature. Available:"VSports" http://www.ncbi.nlm.nih.gov/pubmed/21508958. - PMC - PubMed
-
- Aagaard K, et al (under review) A Comprehensive Strategy for Sampling the Human Microbiome.
-
- Human Microbiome Project Consortium (2012) A framework for human microbiome research. Nature. 2012 Jun 13 486(7402): 215–21 doi: 10.1038/nature11209. - "VSports" PMC - PubMed
-
- Holt RA, Jones SJM (2008) The new paradigm of flow cell sequencing. Genome Res 18: 839–846 doi:10.1101/gr.073262.107. - PubMed
Publication types
- "V体育安卓版" Actions
MeSH terms
- "V体育安卓版" Actions
- "V体育平台登录" Actions
- "VSports最新版本" Actions
- "V体育官网入口" Actions
- Actions (VSports在线直播)
- "V体育ios版" Actions
- "VSports app下载" Actions
- VSports注册入口 - Actions
- V体育平台登录 - Actions
- Actions (V体育平台登录)
- Actions (VSports注册入口)
- "VSports注册入口" Actions
- "V体育官网" Actions
Substances
"VSports在线直播" Grants and funding
LinkOut - more resources
"VSports" Full Text Sources
"V体育官网入口" Other Literature Sources

