Asymmetrically modified nucleosomes
- PMID: 23021224
- PMCID: PMC3498816
- DOI: 10.1016/j.cell.2012.09.002 (VSports最新版本)
Asymmetrically modified nucleosomes
Abstract
Mononucleosomes, the basic building blocks of chromatin, contain two copies of each core histone VSports手机版. The associated posttranslational modifications regulate essential chromatin-dependent processes, yet whether each histone copy is identically modified in vivo is unclear. We demonstrate that nucleosomes in embryonic stem cells, fibroblasts, and cancer cells exist in both symmetrically and asymmetrically modified populations for histone H3 lysine 27 di/trimethylation (H3K27me2/3) and H4K20me1. Further, we obtained direct physical evidence for bivalent nucleosomes carrying H3K4me3 or H3K36me3 along with H3K27me3, albeit on opposite H3 tails. Bivalency at target genes was resolved upon differentiation of ES cells. Polycomb repressive complex 2-mediated methylation of H3K27 was inhibited when nucleosomes contain symmetrically, but not asymmetrically, placed H3K4me3 or H3K36me3. These findings uncover a potential mechanism for the incorporation of bivalent features into nucleosomes and demonstrate how asymmetry might set the stage to diversify functional nucleosome states. .
Copyright © 2012 Elsevier Inc. All rights reserved. V体育安卓版.
Figures

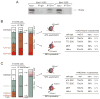
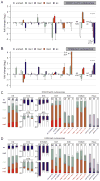
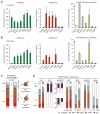

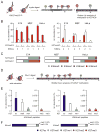

Comment in
-
Chromatin: Histone sibling rivalry.Nat Rev Mol Cell Biol. 2012 Nov;13(11):683. doi: 10.1038/nrm3462. Epub 2012 Oct 17. Nat Rev Mol Cell Biol. 2012. PMID: 23072886 No abstract available.
References (VSports在线直播)
-
- Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem. 2005;280:12065–12068. - PubMed (V体育官网入口)
-
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. - VSports - PMC - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. - PubMed
Publication types
MeSH terms
- VSports手机版 - Actions
- V体育ios版 - Actions
- "V体育平台登录" Actions
- Actions (V体育ios版)
- "V体育平台登录" Actions
Substances
- "V体育ios版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
V体育安卓版 - Other Literature Sources

