Regulation of the tumor suppressor PTEN by SUMO
- PMID: 23013792
- PMCID: PMC3461367
- DOI: 10.1038/cddis.2012.135
Regulation of the tumor suppressor PTEN by SUMO (VSports注册入口)
Abstract
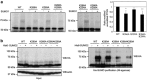
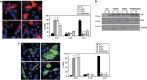
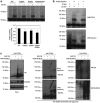
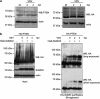
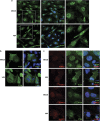
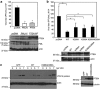
The crucial function of the PTEN tumor suppressor in multiple cellular processes suggests that its activity must be tightly controlled. Both, membrane association and a variety of post-translational modifications, such as acetylation, phosphorylation, and mono- and polyubiquitination, have been reported to regulate PTEN activity. Here, we demonstrated that PTEN is also post-translationally modified by the small ubiquitin-like proteins, small ubiquitin-related modifier 1 (SUMO1) and SUMO2 VSports手机版. We identified lysine residue 266 and the major monoubiquitination site 289, both located within the C2 domain required for PTEN membrane association, as SUMO acceptors in PTEN. We demonstrated the existence of a crosstalk between PTEN SUMOylation and ubiquitination, with PTEN-SUMO1 showing a reduced capacity to form covalent interactions with monoubiquitin and accumulation of PTEN-SUMO2 conjugates after inhibition of the proteasome. Moreover, we found that virus infection induces PTEN SUMOylation and favors PTEN localization at the cell membrane. Finally, we demonstrated that SUMOylation contributes to the control of virus infection by PTEN. .
Figures

"VSports" References
-
- Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. - PubMed
-
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. - PubMed
-
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. - VSports app下载 - PubMed
-
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. - "V体育安卓版" PMC - PubMed
-
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. - "V体育安卓版" PubMed
V体育平台登录 - Publication types
- "V体育官网" Actions
MeSH terms
- "V体育2025版" Actions
- "VSports app下载" Actions
- "VSports注册入口" Actions
- VSports最新版本 - Actions
- VSports在线直播 - Actions
- V体育官网入口 - Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- "V体育官网入口" Actions
- Actions (VSports最新版本)
- "VSports手机版" Actions
- "VSports在线直播" Actions
VSports - Substances
- "V体育安卓版" Actions
- "V体育2025版" Actions
- VSports注册入口 - Actions
LinkOut - more resources
Full Text Sources
V体育安卓版 - Molecular Biology Databases
Research Materials
Miscellaneous

