Induction and molecular signature of pathogenic TH17 cells
- PMID: 22961052
- PMCID: "V体育ios版" PMC3459594
- DOI: 10.1038/ni.2416
Induction and molecular signature of pathogenic TH17 cells
Abstract
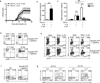
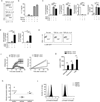
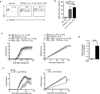
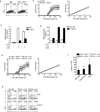
Interleukin 17 (IL-17)-producing helper T cells (T(H)17 cells) are often present at the sites of tissue inflammation in autoimmune diseases, which has led to the conclusion that T(H)17 cells are main drivers of autoimmune tissue injury. However, not all T(H)17 cells are pathogenic; in fact, T(H)17 cells generated with transforming growth factor-β1 (TGF-β1) and IL-6 produce IL-17 but do not readily induce autoimmune disease without further exposure to IL-23. Here we found that the production of TGF-β3 by developing T(H)17 cells was dependent on IL-23, which together with IL-6 induced very pathogenic T(H)17 cells. Moreover, TGF-β3-induced T(H)17 cells were functionally and molecularly distinct from TGF-β1-induced T(H)17 cells and had a molecular signature that defined pathogenic effector T(H)17 cells in autoimmune disease VSports手机版. .
Figures

References (V体育2025版)
-
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. - PubMed
-
- Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. - PubMed
-
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. - PubMed
-
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. - PubMed
-
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. - PubMed
Publication types
- "VSports手机版" Actions
MeSH terms
- Actions (V体育2025版)
- "VSports注册入口" Actions
- VSports app下载 - Actions
- Actions (VSports app下载)
- V体育平台登录 - Actions
- VSports - Actions
Substances (V体育官网)
- V体育官网 - Actions
- "V体育ios版" Actions
- "VSports app下载" Actions
Associated data
- V体育ios版 - Actions
Grants and funding
- V体育平台登录 - K99 AI075285/AI/NIAID NIH HHS/United States
- K01DK090105/DK/NIDDK NIH HHS/United States
- R56 AI093903/AI/NIAID NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- R01 AI093903/AI/NIAID NIH HHS/United States (V体育ios版)
- P01 AI045757/AI/NIAID NIH HHS/United States
- AI075285/AI/NIAID NIH HHS/United States
- R00 AI075285/AI/NIAID NIH HHS/United States
- NS045937/NS/NINDS NIH HHS/United States (V体育官网)
- P01 AI073748/AI/NIAID NIH HHS/United States
- R01 NS030843/NS/NINDS NIH HHS/United States
- NS030843/NS/NINDS NIH HHS/United States
- AI045757/AI/NIAID NIH HHS/United States (V体育平台登录)
- DP1-OD003958-01/OD/NIH HHS/United States
- "VSports app下载" R01 NS045937/NS/NINDS NIH HHS/United States
- R01 AI091568/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 1P01HG005062-01/HG/NHGRI NIH HHS/United States
- K01 DK090105/DK/NIDDK NIH HHS/United States
- R37 NS030843/NS/NINDS NIH HHS/United States
- AI073748/AI/NIAID NIH HHS/United States
- AI093903/AI/NIAID NIH HHS/United States
- R29 NS030843/NS/NINDS NIH HHS/United States
"V体育官网入口" LinkOut - more resources
"VSports最新版本" Full Text Sources
Other Literature Sources
V体育2025版 - Medical
Molecular Biology Databases

