Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability
- PMID: 22904069
- PMCID: PMC3479199
- DOI: 10.1093/nar/gks745
VSports app下载 - Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability
Abstract
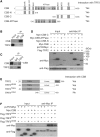
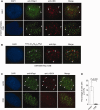
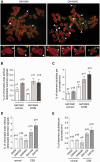
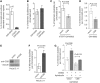
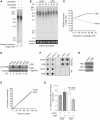
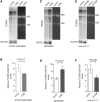
The majority of Cockayne syndrome (CS) patients carry a mutation in Cockayne Syndrome group B (CSB), a large nuclear protein implicated in DNA repair, transcription and chromatin remodeling. However, whether CSB may play a role in telomere metabolism has not yet been characterized. Here, we report that CSB physically interacts with TRF2, a duplex telomeric DNA binding protein essential for telomere protection. We find that CSB localizes at a small subset of human telomeres and that it is required for preventing the formation of telomere dysfunction-induced foci (TIF) in CS cells. We find that CS cells or CSB knockdown cells accumulate telomere doublets, the suppression of which requires CSB. We find that overexpression of CSB in CS cells promotes telomerase-dependent telomere lengthening, a phenotype that is associated with a decrease in the amount of telomere-bound TRF1, a negative mediator of telomere length maintenance. Furthermore, we show that CS cells or CSB knockdown cells exhibit misregulation of TERRA, a large non-coding telomere repeat-containing RNA important for telomere maintenance. Taken together, these results suggest that CSB is required for maintaining the homeostatic level of TERRA, telomere length and integrity. These results further imply that CS patients carrying CSB mutations may be defective in telomere maintenance VSports手机版. .
VSports在线直播 - Figures
References
-
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. - PubMed
-
- Palm W, de Lange T. How shelterin protects Mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. - PubMed
-
- Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. - PubMed
-
- Walker JR, Zhu XD. Post-translational modification of TRF1 and TRF2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012;133:421–434. - PubMed
-
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. - PubMed
Publication types (VSports最新版本)
MeSH terms
- "VSports在线直播" Actions
- "V体育ios版" Actions
- V体育官网入口 - Actions
- Actions (V体育2025版)
- Actions (VSports注册入口)
- "VSports" Actions
- Actions (VSports app下载)
- VSports在线直播 - Actions
Substances
- Actions (VSports在线直播)
- "VSports手机版" Actions
- Actions (V体育官网)
- "V体育平台登录" Actions
- Actions (V体育平台登录)
- VSports手机版 - Actions
LinkOut - more resources
Full Text Sources (V体育安卓版)
Research Materials
Miscellaneous

