HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers
- PMID: 22863008
- PMCID: PMC3438889
- DOI: 10.1016/j.cell.2012.06.031
"V体育平台登录" HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers
Abstract
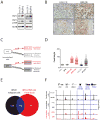
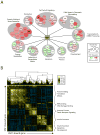
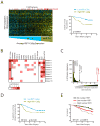
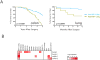
Heat-Shock Factor 1 (HSF1), master regulator of the heat-shock response, facilitates malignant transformation, cancer cell survival, and proliferation in model systems. The common assumption is that these effects are mediated through regulation of heat-shock protein (HSP) expression. However, the transcriptional network that HSF1 coordinates directly in malignancy and its relationship to the heat-shock response have never been defined. By comparing cells with high and low malignant potential alongside their nontransformed counterparts, we identify an HSF1-regulated transcriptional program specific to highly malignant cells and distinct from heat shock VSports手机版. Cancer-specific genes in this program support oncogenic processes: cell-cycle regulation, signaling, metabolism, adhesion and translation. HSP genes are integral to this program, however, many are uniquely regulated in malignancy. This HSF1 cancer program is active in breast, colon and lung tumors isolated directly from human patients and is strongly associated with metastasis and death. Thus, HSF1 rewires the transcriptome in tumorigenesis, with prognostic and therapeutic implications. .
Copyright © 2012 Elsevier Inc. All rights reserved V体育安卓版. .
Figures



References
-
- Anckar J, Sistonen L. Regulation of HSF1 Function in the Heat Stress Response: Implications in Aging and Disease. Annu Rev Biochem. 2011;80:1089–1115. - PubMed
-
- Christians ES, Yan LJ, Benjamin IJ. Heat shock factor 1 and heat shock proteins: Critical partners in protection against acute cell injury. Crit Care Med. 2002;30:S43–S50. - V体育官网入口 - PubMed
Publication types (V体育2025版)
- V体育平台登录 - Actions
MeSH terms
- Actions (V体育2025版)
- Actions (VSports手机版)
- VSports在线直播 - Actions
- Actions (VSports注册入口)
- "V体育官网" Actions
- Actions (V体育安卓版)
- Actions (VSports最新版本)
"V体育2025版" Substances
- V体育官网入口 - Actions
Grants and funding
"VSports手机版" LinkOut - more resources
Full Text Sources
"VSports" Other Literature Sources
Molecular Biology Databases

