V体育官网 - Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development
- PMID: 22858678
- PMCID: PMC3551862 (V体育平台登录)
- DOI: 10.4161/rna.21089
Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development (VSports最新版本)
Abstract
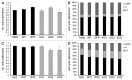
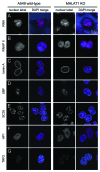
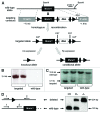
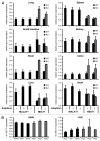
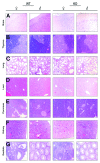
The metastasis-associated lung adenocarcinoma transcript 1, MALAT1, is a long non-coding RNA (lncRNA) that has been discovered as a marker for lung cancer metastasis VSports手机版. It is highly abundant, its expression is strongly regulated in many tumor entities including lung adenocarcinoma and hepatocellular carcinoma as well as physiological processes, and it is associated with many RNA binding proteins and highly conserved throughout evolution. The nuclear transcript MALAT-1 has been functionally associated with gene regulation and alternative splicing and its regulation has been shown to impact proliferation, apoptosis, migration and invasion. Here, we have developed a human and a mouse knockout system to study the loss-of-function phenotypes of this important ncRNA. In human tumor cells, MALAT1 expression was abrogated using Zinc Finger Nucleases. Unexpectedly, the quantitative loss of MALAT1 did neither affect proliferation nor cell cycle progression nor nuclear architecture in human lung or liver cancer cells. Moreover, genetic loss of Malat1 in a knockout mouse model did not give rise to any obvious phenotype or histological abnormalities in Malat1-null compared with wild-type animals. Thus, loss of the abundant nuclear long ncRNA MALAT1 is compatible with cell viability and normal development. .
Figures

"V体育安卓版" References
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
-
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. - "V体育2025版" DOI - PubMed
Publication types
"V体育官网入口" MeSH terms
- Actions (VSports注册入口)
- Actions (V体育安卓版)
- Actions (VSports手机版)
- Actions (V体育官网)
- "VSports app下载" Actions
- "V体育安卓版" Actions
- V体育官网入口 - Actions
Substances (V体育ios版)
- Actions (VSports)
- "VSports在线直播" Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
