5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription
- PMID: 22820989
- PMCID: PMC3414690
- DOI: 10.1038/nsmb.2346
5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription (VSports最新版本)
Abstract
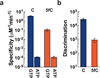
Although the roles of 5-methylcytosine and 5-hydroxymethylcytosine in epigenetic regulation of gene expression are well established, the functional effects of 5-formylcytosine and 5-carboxylcytosine on the process of transcription are not clear. Here we report a systematic study of the effects of five different forms of cytosine in DNA on mammalian and yeast RNA polymerase II transcription, providing new insights into potential functional interplay between cytosine methylation status and transcription VSports手机版. .
Figures

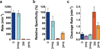
Comment in (VSports app下载)
-
New functions for DNA modifications by TET-JBP.Nat Struct Mol Biol. 2012 Nov;19(11):1061-4. doi: 10.1038/nsmb.2437. Nat Struct Mol Biol. 2012. PMID: 23132381 Free PMC article.
References
-
- Jaenisch R, Bird A. Nat Genet. 2003;(33 Suppl):245–254. - PubMed
-
- Law JA, Jacobsen SE. Nat Rev Genet. 2010;11:204–220. - PMC (VSports app下载) - PubMed
-
- Pfaffeneder T, et al. Angew Chem Int Ed Engl. 2011;50:7008–7012. - PubMed
Publication types
- "V体育官网" Actions
MeSH terms
- Actions (V体育官网入口)
- Actions (V体育ios版)
- Actions (V体育2025版)
- "VSports最新版本" Actions
- Actions (V体育官网入口)
- "VSports app下载" Actions
- VSports注册入口 - Actions
- VSports在线直播 - Actions
- "V体育安卓版" Actions
Substances
- Actions (V体育ios版)
- Actions (V体育平台登录)
- V体育2025版 - Actions
- V体育安卓版 - Actions
- Actions (V体育ios版)
Grants and funding
LinkOut - more resources
VSports手机版 - Full Text Sources
Other Literature Sources
Molecular Biology Databases

