AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation
- PMID: 22772155
- PMCID: PMC3427411
- DOI: 10.1038/nchembio.1042
AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation
VSports - Abstract
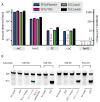
Activation-induced deaminase (AID)/APOBEC-family cytosine deaminases, known to function in diverse cellular processes from antibody diversification to mRNA editing, have also been implicated in DNA demethylation, a major process for transcriptional activation. Although oxidation-dependent pathways for demethylation have been described, pathways involving deamination of either 5-methylcytosine (5mC) or 5-hydroxymethylcytosine (5hmC) have emerged as alternatives. Here we address the biochemical plausibility of deamination-coupled demethylation. We found that purified AID/APOBECs have substantially reduced activity on 5mC relative to cytosine, their canonical substrate, and no detectable deamination of 5hmC. This finding was explained by the reactivity of a series of modified substrates, where steric bulk was increasingly detrimental to deamination. Further, upon AID/APOBEC overexpression, the deamination product of 5hmC was undetectable in genomic DNA, whereas oxidation intermediates remained detectable. Our results indicate that the steric requirements for cytosine deamination are one intrinsic barrier to the proposed function of deaminases in DNA demethylation VSports手机版. .
Conflict of interest statement
The authors declare no competing financial interests.
Figures
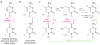
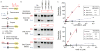

References
-
- Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv Immunol. 2007;94:215–244. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed (VSports注册入口)
Publication types
- VSports app下载 - Actions
MeSH terms
- Actions (VSports最新版本)
- "V体育官网入口" Actions
- "V体育官网" Actions
- "V体育官网" Actions
Substances
- V体育官网 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
VSports在线直播 - Molecular Biology Databases

