VSports - Methylation markers of early-stage non-small cell lung cancer
- PMID: 22768131
- PMCID: PMC3387223
- DOI: 10.1371/journal.pone.0039813 (VSports)
"V体育安卓版" Methylation markers of early-stage non-small cell lung cancer
Erratum in
-
Correction: Methylation Markers of Early-Stage Non-Small Cell Lung Cancer.PLoS One. 2017 Jan 20;12(1):e0170833. doi: 10.1371/journal.pone.0170833. eCollection 2017. PLoS One. 2017. PMID: 28107536 Free PMC article.
"V体育官网入口" Abstract
Background: Despite of intense research in early cancer detection, there is a lack of biomarkers for the reliable detection of malignant tumors, including non-small cell lung cancer (NSCLC). DNA methylation changes are common and relatively stable in various types of cancers, and may be used as diagnostic or prognostic biomarkers. VSports手机版.
Methods: We performed DNA methylation profiling of samples from 48 patients with stage I NSCLC and 18 matching cancer-free lung samples using microarrays that cover the promoter regions of more than 14,500 genes. We correlated DNA methylation changes with gene expression levels and performed survival analysis. V体育安卓版.
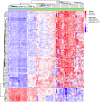
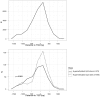
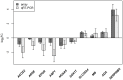
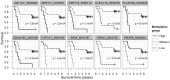
Results: We observed hypermethylation of 496 CpGs in 379 genes and hypomethylation of 373 CpGs in 335 genes in NSCLC. Compared to adenocarcinoma samples, squamous cell carcinoma samples had 263 CpGs in 223 hypermethylated genes and 513 CpGs in 436 hypomethylated genes. 378 of 869 (43. 5%) CpG sites discriminating the NSCLC and control samples showed an inverse correlation between CpG site methylation and gene expression levels. As a result of a survival analysis, we found 10 CpGs in 10 genes, in which the methylation level differs in different survival groups. V体育ios版.
Conclusions: We have identified a set of genes with altered methylation in NSCLC and found that a minority of them showed an inverse correlation with gene expression levels VSports最新版本. We also found a set of genes that associated with the survival of the patients. These newly-identified marker candidates for the molecular screening of NSCLC will need further analysis in order to determine their clinical utility. .
Conflict of interest statement
Figures
References
-
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. - PubMed
-
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. - PubMed
-
- Uribe-Lewis S, Woodfine K, Stojic L, Murrell A. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev Mol Med. 2011;13:e2. - PubMed
-
- Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059–1068. - PubMed
V体育官网 - Publication types
VSports手机版 - MeSH terms
- Actions (VSports手机版)
- VSports注册入口 - Actions
- "VSports最新版本" Actions
- "VSports" Actions
- VSports最新版本 - Actions
- Actions (V体育2025版)
- "VSports app下载" Actions
- V体育2025版 - Actions
- "VSports" Actions
- "V体育官网入口" Actions
- "V体育安卓版" Actions
- Actions (VSports最新版本)
- "VSports" Actions
- "V体育ios版" Actions
- V体育安卓版 - Actions
- V体育2025版 - Actions
"V体育安卓版" Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
V体育2025版 - Research Materials

