Activated fluid transport regulates bacterial-epithelial interactions and significantly shifts the murine colonic microbiome
- PMID: 22614705
- PMCID: PMC3427217
- DOI: 10.4161/gmic.20529
Activated fluid transport regulates bacterial-epithelial interactions and significantly shifts the murine colonic microbiome (V体育安卓版)
Abstract
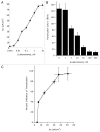
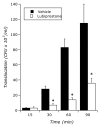
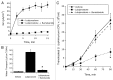
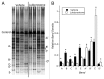
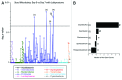
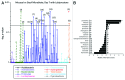
Within the intestinal mucosa, epithelial cells serve multiple functions to partition the lumen from the lamina propria. As part of their natural function, intestinal epithelial cells actively transport electrolytes with passive water movement as a mechanism for mucosal hydration. Here, we hypothesized that electrogenic Cl(-) secretion, and associated mucosal hydration, influences bacterial-epithelial interactions and significantly influences the composition of the intestinal microbiota. An initial screen of different epithelial secretagogues identified lubiprostone as the most potent agonist for which to define these principles. In in vitro studies using cultured T84 cells, lubiprostone decreased E VSports手机版. coli translocation in a concentration-dependent manner (p < 0. 001) and decreased S. typhimurium internalization and translocation by as much as 71 ± 6% (p < 0. 01). Such decreases in bacterial translocation were abolished by inhibition of electrogenic Cl(-) secretion and water transport using the Na/K/Cl(-) antagonist bumetanide (p < 0. 01). Extensions of these findings to microbiome analysis in vivo revealed that lubiprostone delivered orally to mice fundamentally shifted the intestinal microbiota, with notable changes within the Firmicutes and Bacteroidetes phyla of resident colonic bacteria. Such findings document a previously unappreciated role for epithelial Cl(-) secretion and water transport in influencing bacterial-epithelial interactions and suggest that active mucosal hydration functions as a primitive innate epithelial defense mechanism. .
Figures


References
-
- Barrett KE, Keely SJ. Chloride secretion by the intestinal epithelium: molecular basis and regulatory aspects. Annu Rev Physiol. 2000;62:535–72. doi: 10.1146/annurev.physiol.62.1.535. - DOI (VSports注册入口) - PubMed
VSports - Publication types
- Actions (VSports app下载)
MeSH terms
- V体育ios版 - Actions
- V体育平台登录 - Actions
- V体育2025版 - Actions
- VSports最新版本 - Actions
- "V体育安卓版" Actions
- Actions (V体育官网入口)
- "V体育ios版" Actions
- V体育2025版 - Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- "VSports app下载" Actions
- Actions (V体育2025版)
Substances (V体育平台登录)
- VSports app下载 - Actions
- V体育平台登录 - Actions
"V体育安卓版" Grants and funding
LinkOut - more resources
VSports在线直播 - Full Text Sources
Research Materials
