"VSports在线直播" Target specificity of an autoreactive pathogenic human γδ-T cell receptor in myositis
- PMID: 22549773
- PMCID: "VSports在线直播" PMC3375522
- DOI: V体育安卓版 - 10.1074/jbc.M112.356709
Target specificity of an autoreactive pathogenic human γδ-T cell receptor in myositis
Abstract
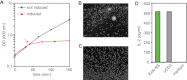
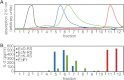
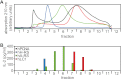
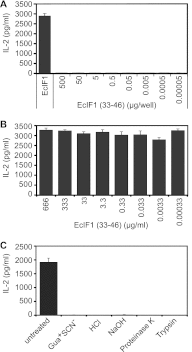
In polymyositis and inclusion body myositis, muscle fibers are surrounded and invaded by CD8-positive cytotoxic T cells expressing the αβ-T cell receptor (αβ-TCR) for antigen. In a rare variant of myositis, muscle fibers are similarly attacked by CD8-negative T cells expressing the γδ-TCR (γδ-T cell-mediated myositis). We investigated the antigen specificity of a human γδ-TCR previously identified in an autoimmune tissue lesion of γδ-T cell-mediated myositis. We show that this Vγ1. 3Vδ2-TCR, termed M88, recognizes various proteins from different species. Several of these proteins belong to the translational apparatus, including some bacterial and human aminoacyl-tRNA synthetases (AA-RS). Specifically, M88 recognizes histidyl-tRNA synthetase, an antigen known to be also targeted by autoantibodies called anti-Jo-1 VSports手机版. The M88 target epitope is strictly conformational, independent of post-translational modification, and exposed on the surface of the respective antigenic protein. Extensive mutagenesis of the translation initiation factor-1 from Escherichia coli (EcIF1), which served as a paradigm antigen with known structure, showed that a short α-helical loop around amino acids 39 to 42 of EcIF1 is a major part of the M88 epitope. Mutagenesis of M88 showed that the complementarity determining regions 3 of both γδ-TCR chains contribute to antigen recognition. M88 is the only known example of a molecularly characterized γδ-TCR expressed by autoaggressive T cells in tissue. The observation that AA-RS are targeted by a γδ-T cell and by autoantibodies reveals an unexpected link between T cell and antibody responses in autoimmune myositis. .
Figures


"V体育安卓版" References
-
- Chien Y. H., Konigshofer Y. (2007) Antigen recognition by γδ T cells. Immunol. Rev. 215, 46–58 - PubMed
-
- Beetz S., Wesch D., Marischen L., Welte S., Oberg H. H., Kabelitz D. (2008) Innate immune functions of human γδ T cells. Immunobiology 213, 173–182 - V体育2025版 - PubMed
-
- Hayday A. C. (2009) γδ T cells and the lymphoid stress-surveillance response. Immunity 31, 184–196 - V体育官网 - PubMed
-
- Bonneville M., O'Brien R. L., Born W. K. (2010) γδ T cell effector functions: A blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 - PubMed
Publication types
- Actions (VSports手机版)
V体育官网入口 - MeSH terms
- Actions (VSports手机版)
- V体育安卓版 - Actions
- Actions (V体育ios版)
- Actions (V体育安卓版)
- "V体育官网入口" Actions
- Actions (VSports)
- Actions (V体育官网)
- "V体育ios版" Actions
- VSports - Actions
- V体育官网入口 - Actions
- Actions (VSports最新版本)
- Actions (V体育安卓版)
Substances
- Actions (V体育安卓版)
- V体育2025版 - Actions
- VSports app下载 - Actions
"VSports手机版" LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

