MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition
- PMID: 22430491
- PMCID: PMC3328367
- DOI: 10.1084/jem.20111512
MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition
Abstract
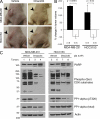
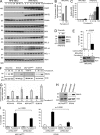
Estrogen, progesterone, and HER2 receptor-negative triple-negative breast cancers encompass the most clinically challenging subtype for which targeted therapeutics are lacking VSports手机版. We find that triple-negative tumors exhibit elevated MYC expression, as well as altered expression of MYC regulatory genes, resulting in increased activity of the MYC pathway. In primary breast tumors, MYC signaling did not predict response to neoadjuvant chemotherapy but was associated with poor prognosis. We exploit the increased MYC expression found in triple-negative breast cancers by using a synthetic-lethal approach dependent on cyclin-dependent kinase (CDK) inhibition. CDK inhibition effectively induced tumor regression in triple-negative tumor xenografts. The proapoptotic BCL-2 family member BIM is up-regulated after CDK inhibition and contributes to this synthetic-lethal mechanism. These results indicate that aggressive breast tumors with elevated MYC are uniquely sensitive to CDK inhibitors. .
Figures


"VSports注册入口" References
-
- Alles M.C., Gardiner-Garden M., Nott D.J., Wang Y., Foekens J.A., Sutherland R.L., Musgrove E.A., Ormandy C.J. 2009. Meta-analysis and gene set enrichment relative to er status reveal elevated activity of MYC and E2F in the “basal” breast cancer subgroup. PLoS ONE. 4:e4710 10.1371/journal.pone.0004710 - DOI - PMC - PubMed
-
- Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. 2007. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 109:1721–1728. 10.1002/cncr.22618 - DOI - PubMed
Publication types
MeSH terms
- VSports app下载 - Actions
- Actions (V体育安卓版)
- VSports - Actions
- "VSports手机版" Actions
- VSports注册入口 - Actions
- VSports最新版本 - Actions
- "V体育平台登录" Actions
- "V体育2025版" Actions
- "V体育ios版" Actions
- "VSports最新版本" Actions
- "V体育平台登录" Actions
- "V体育官网" Actions
- VSports在线直播 - Actions
Substances
- VSports手机版 - Actions
- "VSports手机版" Actions
- "V体育ios版" Actions
- "VSports app下载" Actions
- Actions (V体育平台登录)
- VSports最新版本 - Actions
- V体育ios版 - Actions
- Actions (V体育官网)
Grants and funding
- 1R01CA136717/CA/NCI NIH HHS/United States
- 1K23CA121994/CA/NCI NIH HHS/United States
- CA58207/CA/NCI NIH HHS/United States
- U01 CA080098/CA/NCI NIH HHS/United States
- CA3360/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- R01 CA136717/CA/NCI NIH HHS/United States
- V体育官网 - P50 CA058207/CA/NCI NIH HHS/United States
- 2T32CA108462/CA/NCI NIH HHS/United States
- U01 CA079778/CA/NCI NIH HHS/United States
- U10 CA080098/CA/NCI NIH HHS/United States
- U01CA080098/CA/NCI NIH HHS/United States
- CA31964/CA/NCI NIH HHS/United States
- 1K08CA104032/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- V体育官网 - K23 CA121994/CA/NCI NIH HHS/United States
- V体育ios版 - K08 CA104032/CA/NCI NIH HHS/United States
- VSports app下载 - 5P50CA058207/CA/NCI NIH HHS/United States
- T32 CA108462/CA/NCI NIH HHS/United States
- U01CA079778/CA/NCI NIH HHS/United States (VSports在线直播)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous (V体育安卓版)

